4F1N
 
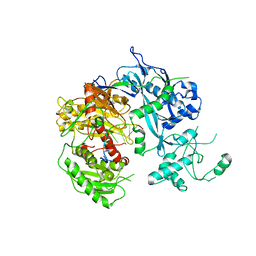 | | Crystal structure of Kluyveromyces polysporus Argonaute with a guide RNA | | Descriptor: | KpAGO, RNA 5'-R(P*UP*AP*AP*AP*AP*AP*AP*AP*A)-3' | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure of yeast Argonaute with guide RNA.
Nature, 486, 2012
|
|
1YWG
 
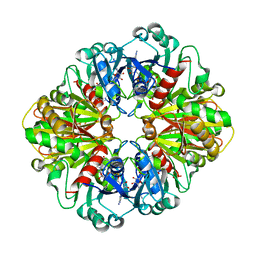 | | The structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Satchell, J.F, Klonis, N, Malby, R.L, Adisa, A, Alpyurek, A.E, Colman, P.M, Tilley, L. | | Deposit date: | 2005-02-17 | | Release date: | 2005-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1R3Q
 
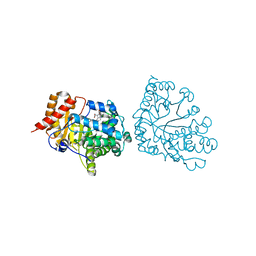 | | Uroporphyrinogen Decarboxylase in complex with coproporphyrinogen-I | | Descriptor: | CARBON DIOXIDE, COPROPORPHYRINOGEN I, Uroporphyrinogen Decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Kushner, J.P, Hill, C.P. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase
Embo J., 22, 2003
|
|
4F2J
 
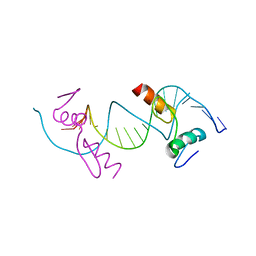 | | Crystal structure of ZNF217 bound to DNA, P6522 crystal form | | Descriptor: | 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', ZINC ION, Zinc finger protein 217 | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2012-05-08 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217
J.Biol.Chem., 288, 2013
|
|
7W3W
 
 | | X-ray structure of apo-VmFbpA, a ferric ion-binding protein from Vibrio metschnikovii | | Descriptor: | Iron-utilization periplasmic protein | | Authors: | Lu, P, Sui, M, Zhang, M, Nagata, K. | | Deposit date: | 2021-11-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Rosmarinic Acid and Sodium Citrate Have a Synergistic Bacteriostatic Effect against Vibrio Species by Inhibiting Iron Uptake.
Int J Mol Sci, 22, 2021
|
|
1DGC
 
 | |
1JNZ
 
 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, SULFITE ION, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M, Ermler, U. | | Deposit date: | 2001-07-26 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6EH4
 
 | |
2LG3
 
 | | Structure of the duplex containing HNE derived (6S,8R,11S) gamma-HO-PdG when placed opposite dT | | Descriptor: | (4S)-nonane-1,4-diol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*GP*CP*TP*AP*GP*C)-3') | | Authors: | Huang, H, Wang, H, Kozekova, A, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ring-chain tautomerization of trans-4-hydroxynonenal derived (6S,8R,11S) gamma-hydroxy-1,N2-propano-deoxyguanosine adduct when placed opposite 2'-deoxythymidine in duplex
To be Published
|
|
8WCM
 
 | |
1JPU
 
 | | Crystal Structure of Bacillus Stearothermophilus Glycerol Dehydrogenase | | Descriptor: | ZINC ION, glycerol dehydrogenase | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
4B9H
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
1V3E
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with ZANAMAVIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
3GVP
 
 | | Human SAHH-like domain of human adenosylhomocysteinase 3 | | Descriptor: | Adenosylhomocysteinase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Siponen, M.I, Wisniewska, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schueler, H. | | Deposit date: | 2009-03-31 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human S-adenosyl homocysteine hydrolase-like 2 protein crystal structure
To be Published
|
|
8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
2O7B
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides, complexed with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
3TK3
 
 | | Cytochrome P450 2B4 mutant L437A in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Jang, H.H, Wilderman, P.R, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme.
Febs J., 279, 2012
|
|
6UZV
 
 | | The structure of a red shifted photosystem I complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Toporik, H, Williams, D, Chiu, P.L, Mazor, Y. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of a red-shifted photosystem I reveals a red site in the core antenna.
Nat Commun, 11, 2020
|
|
8FH3
 
 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Tubby-related protein 3, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
8FGW
 
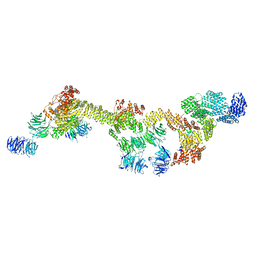 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
4BI9
 
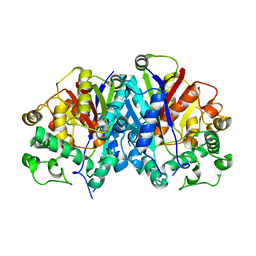 | | Crystal structure of wild-type SCP2 thiolase from Trypanosoma brucei. | | Descriptor: | 3-KETOACYL-COA THIOLASE, PUTATIVE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
6ECZ
 
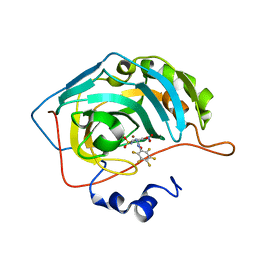 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | Carbonic anhydrase 2, N-[2-hydroxy-3-nitro-5-(nitrosulfonyl)phenyl]-N'-(pentafluorophenyl)urea, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
4BKQ
 
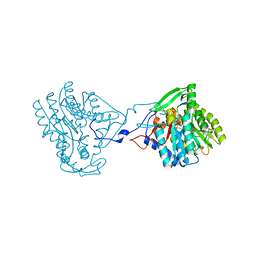 | | Enoyl-ACP reductase from Yersinia pestis (wildtype)with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPZ3_3519 | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
1BLA
 
 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
8FPT
 
 | | STRUCTURE OF ALPHA-SYNUCLEIN FIBRILS DERIVED FROM HUMAN LEWY BODY DEMENTIA TISSUE | | Descriptor: | Alpha-synuclein | | Authors: | Barclay, A.M, Dhavale, D.D, Borcik, C.G, Rau, M.J, Basore, K, Milchberg, M.H, Warmuth, O.A, Kotzbauer, P.T, Rienstra, C.M, Schwieters, C.D. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of alpha-synuclein fibrils derived from human Lewy body dementia tissue.
Biorxiv, 2023
|
|
