5JGZ
 
 | | Spin-Labeled T4 Lysozyme Construct T151V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5CSO
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleoside, cytidine at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5LUU
 
 | | Structure of the first bromodomain of BRD4 with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
6B0H
 
 | | Crystal structure of Pfs25 in complex with the transmission blocking antibody 1262 | | Descriptor: | 1,2-ETHANEDIOL, 1262 antibody, heavy chain, ... | | Authors: | McLeod, B, Scally, S.W, Bosch, A, King, C.R, Julien, J.P. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular definition of multiple sites of antibody inhibition of malaria transmission-blocking vaccine antigen Pfs25.
Nat Commun, 8, 2017
|
|
5CQZ
 
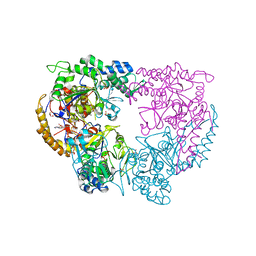 | |
4ADV
 
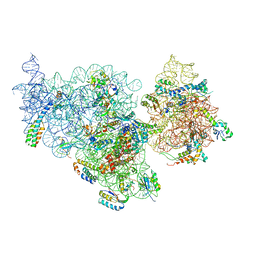 | | Structure of the E. coli methyltransferase KsgA bound to the E. coli 30S ribosomal subunit | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Boehringer, D, O'Farrell, H.C, Rife, J.P, Ban, N. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structural Insights Into Methyltransferase Ksga Function in 30S Ribosomal Subunit Biogenesis
J.Biol.Chem., 287, 2012
|
|
2WZM
 
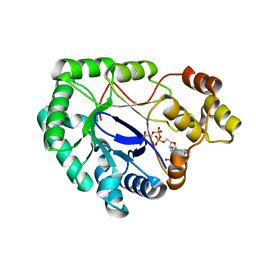 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
5CST
 
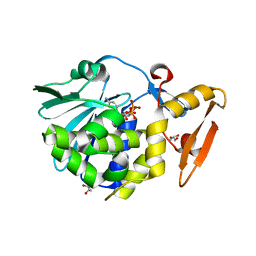 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine diphosphate at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5JNY
 
 | | Crystal Structure of 10E8 Fab | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, CHLORIDE ION, ... | | Authors: | Ofek, G, Kwong, P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.
Plos One, 11, 2016
|
|
6HV2
 
 | | MMP-13 in complex with the peptide IMISF | | Descriptor: | CALCIUM ION, Collagenase 3, GLYCEROL, ... | | Authors: | Mittl, P, Riedl, R, Hohl, D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Drug Design Inspired by Nature: Crystallographic Detection of an Auto-Tailored Protease Inhibitor Template.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
2YPL
 
 | | Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms | | Descriptor: | AGA T-CELL RECEPTOR ALPHA CHAIN, AGA T-CELL RECEPTOR BETA CHAIN, BETA-2-MICROGLOBULIN, ... | | Authors: | Stewart-Jones, G.B, Simpson, P, van der Merwe, P.A, Easterbrook, P, McMichael, A.J, Rowland-Jones, S.L, Jones, E.Y, Gillespie, G.M. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WRB
 
 | | the structure of influenza H2 human singapore hemagglutinin with avian receptor | | Descriptor: | HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid | | Authors: | Liu, J, Stevens, D.J, Haire, L.F, Walker, P.A, Coombs, P.J, Russell, R.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5M1W
 
 | | Structure of a stable G-hairpin | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3') | | Authors: | Gajarsky, M, Zivkovic, M.L, Stadlbauer, P, Pagano, B, Fiala, R, Amato, J, Tomaska, L, Sponer, J, Plavec, J, Trantirek, L. | | Deposit date: | 2016-10-11 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable G-Hairpin.
J. Am. Chem. Soc., 139, 2017
|
|
6YTC
 
 | |
5IU4
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with ZM241385 at 1.7A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6ZQT
 
 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427H/Q433L/K440R) in complex with RalB-GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
6HVL
 
 | | CdaA complex with c-di-AMP and AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
8Q9U
 
 | |
6ZRV
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) in Complex with an Inhibitory Nanobody (VHH-s-a93, Nb93) | | Descriptor: | Plasminogen activator inhibitor 1, VHH-s-a93 (Nb93) | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.
Int J Mol Sci, 21, 2020
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
8BBB
 
 | |
8BBD
 
 | |
7Y5V
 
 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
