1AGO
 
 | |
1X3G
 
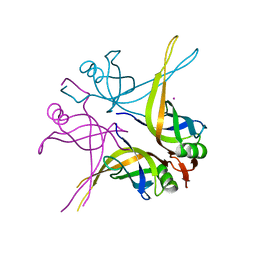 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2AVW
 
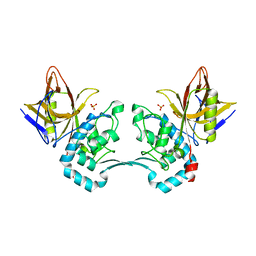 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | Descriptor: | GLYCEROL, IgG-degrading protease, SULFATE ION | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
1NF1
 
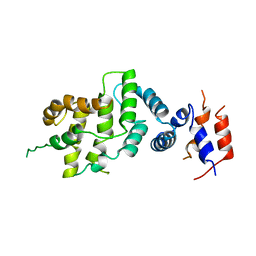 | | THE GAP RELATED DOMAIN OF NEUROFIBROMIN | | Descriptor: | PROTEIN (NEUROFIBROMIN) | | Authors: | Scheffzek, K, Ahmadian, M.R, Wiesmueller, L, Kabsch, W, Stege, P, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1998-07-08 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the GAP-related domain from neurofibromin and its implications.
EMBO J., 17, 1998
|
|
150D
 
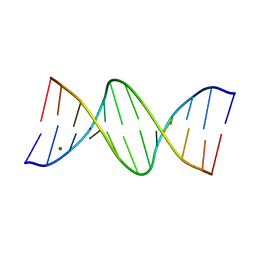 | | GUANINE.1,N6-ETHENOADENINE BASE-PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(EDA)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDA)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Gibson, N.J, Brown, T, Watson, W.P, Hunter, W.N. | | Deposit date: | 1993-12-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine-1,N6-ethenoadenine base pairs in the crystal structure of d(CGCGAATT(epsilon dA)GCG).
Biochemistry, 33, 1994
|
|
165D
 
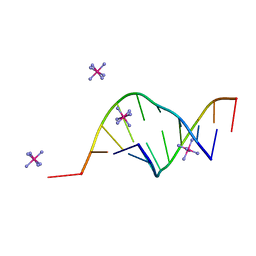 | | THE STRUCTURE OF A MISPAIRED RNA DOUBLE HELIX AT 1.6 ANGSTROMS RESOLUTION AND IMPLICATIONS FOR THE PREDICTION OF RNA SECONDARY STRUCTURE | | Descriptor: | DNA/RNA (5'-R(*GP*CP*UP*UP*CP*GP*GP*CP*)-D(*(BRU))-3'), RHODIUM HEXAMINE ION | | Authors: | Cruse, W, Saludjian, P, Biala, E, Strazewski, P, Prange, T, Kennard, O. | | Deposit date: | 1994-03-21 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a mispaired RNA double helix at 1.6-A resolution and implications for the prediction of RNA secondary structure.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
4GI5
 
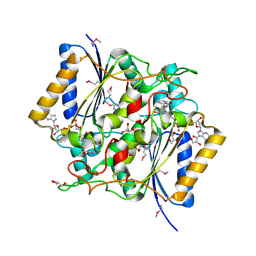 | | Crystal Structure Of a Putative quinone reductase from Klebsiella pneumoniae (Target PSI-013613) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a quinone reductase from Klebsiella pneumoniae with bound FAD
to be published
|
|
1N8J
 
 | |
2QBX
 
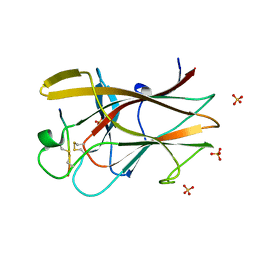 | | EphB2/SNEW Antagonistic Peptide Complex | | Descriptor: | Ephrin type-B receptor 2, SULFATE ION, antagonistic peptide | | Authors: | Chrencik, J.E, Brooun, A, Recht, M.I, Nicola, G, Pasquale, E.B, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-06-18 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the EphB2 receptor in complex with an antagonistic peptide reveals a novel mode of inhibition.
J.Biol.Chem., 282, 2007
|
|
1WCB
 
 | |
3UXE
 
 | | Design, Synthesis and Biological Evaluation of Potent Quinoline and Pyrroloquinoline Ammosamide Analogues as Inhibitors for Quinone Reductase 2 | | Descriptor: | 8-amino-7-chloro-1-methyl-6-(methylideneamino)-2-oxo-1,2-dihydropyrrolo[4,3,2-de]quinoline-4-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Reddy, N.P, Jensen, K.C, Mesecar, A.D, Fanwick, P.E, Cushman, M. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of potent quinoline and pyrroloquinoline ammosamide analogues as inhibitors of quinone reductase 2.
J.Med.Chem., 55, 2012
|
|
1ND7
 
 | | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase | | Descriptor: | WW domain-containing protein 1 | | Authors: | Verdecia, M.A, Joaziero, C.A.P, Wells, N.J, Ferrer, J.-L, Bowman, M.E, Hunter, T, Noel, J.P. | | Deposit date: | 2002-12-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase
Mol.Cell, 11, 2003
|
|
2KW6
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 1 (CDK2-associated protein 1; oral cancer suppressor Deleted in oral cancer 1, DOC-1) from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3057H | | Descriptor: | Cyclin-dependent kinase 2-associated protein 1 | | Authors: | Ertekin, A, Aramini, J.M, Rossi, P, Lee, A.B, Jiang, M, Ciccosanti, C.T, Xiao, R, Swapna, G.V.T, Rost, B, Everett, J.K, Acton, T.B, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Human cyclin-dependent kinase 2-associated protein 1 (CDK2AP1) is dimeric in its disulfide-reduced state, with natively disordered N-terminal region.
J.Biol.Chem., 287, 2012
|
|
1NJQ
 
 | | NMR structure of the single QALGGH zinc finger domain from Arabidopsis thaliana SUPERMAN protein | | Descriptor: | ZINC ION, superman protein | | Authors: | Isernia, C, Bucci, E, Leone, M, Zaccaro, L, Di Lello, P, Digilio, G, Esposito, S, Saviano, M, Di Blasio, B, Pedone, C, Pedone, P.V, Fattorusso, R. | | Deposit date: | 2003-01-02 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Single QALGGH Zinc Finger Domain from the Arabidopsis thaliana SUPERMAN Protein.
Chembiochem, 4, 2003
|
|
2R2D
 
 | | Structure of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, PHOSPHATE ION, ZINC ION, ... | | Authors: | Liu, D, Thomas, P.W, Momb, J, Hoang, Q, Petsko, G.A, Ringe, D, Fast, W. | | Deposit date: | 2007-08-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens.
Biochemistry, 46, 2007
|
|
2R0D
 
 | | Crystal Structure of Autoinhibited Form of Grp1 Arf GTPase Exchange Factor | | Descriptor: | Cytohesin-3, DI(HYDROXYETHYL)ETHER, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | DiNitto, J.P, Delprato, A, Gabe Lee, M.T, Cronin, T.C, Huang, S, Guilherme, A, Czech, M.P, Lambright, D.G. | | Deposit date: | 2007-08-18 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis and Mechanism of Autoregulation in 3-Phosphoinositide-Dependent Grp1 Family Arf GTPase Exchange Factors.
Mol.Cell, 28, 2007
|
|
2R3C
 
 | | Structure of the gp41 N-peptide in complex with the HIV entry inhibitor PIE1 | | Descriptor: | CHLORIDE ION, HIV entry inhibitor PIE1, YTTRIUM (III) ION, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1YXP
 
 | | HIV-1 DIS RNA subtype F- Zn soaked | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', ZINC ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2005-02-22 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
2R24
 
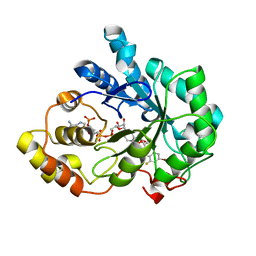 | | Human Aldose Reductase structure | | Descriptor: | Aldose reductase, IDD594, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O.N, Cousido-Siah, A, Haertlein, M, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-24 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.752 Å), X-RAY DIFFRACTION | | Cite: | Quantum model of catalysis based on mobile proton revealed by subatomic X-Ray and neutron diffraction studies of h-Aldose Reductase
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1YZ9
 
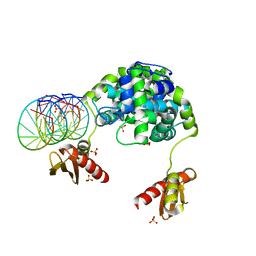 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
3CSK
 
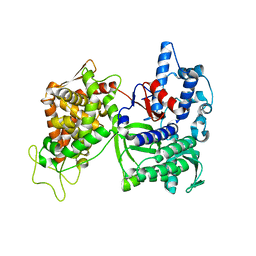 | | Structure of DPP III from Saccharomyces cerevisiae | | Descriptor: | MAGNESIUM ION, Probable dipeptidyl-peptidase 3, ZINC ION | | Authors: | Baral, P.K, Jajcanin, N, Deller, S, Macheroux, P, Abramic, M, Gruber, K. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The first structure of dipeptidyl-peptidase III provides insight into the catalytic mechanism and mode of substrate binding.
J.Biol.Chem., 283, 2008
|
|
3CY5
 
 | |
2KXG
 
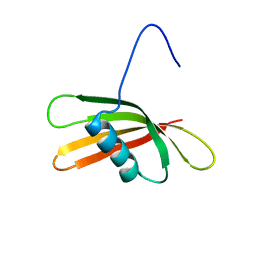 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
1NI2
 
 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
1NIJ
 
 | | YJIA PROTEIN | | Descriptor: | Hypothetical protein yjiA | | Authors: | Khil, P.P, Obmolova, G, Teplyakov, A, Howard, A.J, Gilliland, G.L, Camerini-Otero, R.D, Structure 2 Function Project (S2F) | | Deposit date: | 2002-12-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function.
Proteins, 54, 2004
|
|
