4IGS
 
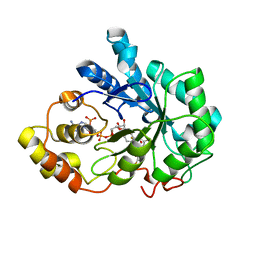 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3SVL
 
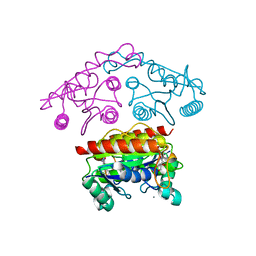 | | Structural basis of the improvement of ChrR - a multi-purpose enzyme | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, protein yieF | | Authors: | Poulain, S, Eswaramoorthy, S, Hienerwadel, R, Bremond, N, Sylvester, M.D, Zhang, Y.B, Van Der Lelie, D, Berthomieu, C, Matin, A.C. | | Deposit date: | 2011-07-12 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of ChrR-A Quinone Reductase with the Capacity to Reduce Chromate.
Plos One, 7, 2012
|
|
3T4B
 
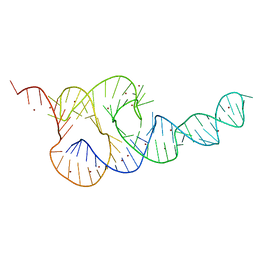 | | Crystal Structure of the HCV IRES pseudoknot domain | | Descriptor: | HCV IRES pseudoknot domain plus crystallization module, NICKEL (II) ION | | Authors: | Berry, K.E, Waghray, S, Mortimer, S.A, Bai, Y, Doudna, J.A. | | Deposit date: | 2011-07-25 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning.
Structure, 19, 2011
|
|
4AVB
 
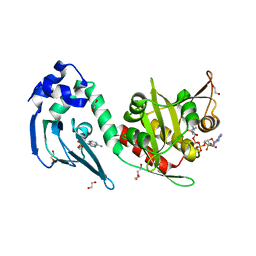 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4CK8
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-(4-(3,4- dichlorophenyl)piperazin-1-yl)phenylcarbamate (LFD) | | Descriptor: | (1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl {4-[4-(3,4-dichlorophenyl)piperazin-1-yl]phenyl}carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4CK9
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFT) | | Descriptor: | (1S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl [4-(propan-2-yl)phenyl]carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
3KYI
 
 | | Crystal structure of the phosphorylated P1 domain of CheA3 in complex with CheY6 from R. sphaeroides | | Descriptor: | CheY6 protein, Putative histidine protein kinase | | Authors: | Bell, C.H, Porter, S.L, Armitage, J.P, Stuart, D.I. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Plos Biol., 8, 2010
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
4CMV
 
 | | Crystal structure of Rv3378c | | Descriptor: | CITRIC ACID, DITERPENE SYNTHASE, MERCURY (II) ION | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CKA
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFS) | | Descriptor: | (1S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-isopropylphenylcarbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4ICC
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WWK
 
 | | Highly stable green fluorescent protein | | Descriptor: | Green fluorescent protein | | Authors: | Sriram, R, George, A, Kesavan, M, Jaimohan, S.M, Kamini, N.R, Easwaramoorthi, S, Ganesh, S, Gunasekaran, K, Ayyadurai, N. | | Deposit date: | 2017-01-02 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Excited State Electronic Interconversion and Structural Transformation of Engineered Red-Emitting Green Fluorescent Protein Mutant.
J.Phys.Chem.B, 123, 2019
|
|
1NYX
 
 | | Ligand binding domain of the human peroxisome proliferator activated receptor gamma in complex with an agonist | | Descriptor: | (2S)-2-ETHOXY-3-{4-[2-(10H-PHENOXAZIN-10-YL)ETHOXY]PHENYL}PROPANOIC ACID, peroxisome proliferator activated receptor gamma | | Authors: | Ebdrup, S, Pettersson, I, Rasmussen, H.B, Deussen, H.-J, Frost Jensen, A, Mortensen, S.B, Fleckner, J, Pridal, L, Nygaard, L, Sauerberg, P. | | Deposit date: | 2003-02-14 | | Release date: | 2003-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and biological and structural characterization of the dual-acting peroxisome proliferator-activated receptor alpha/gamma agonist ragaglitazar
J.MED.CHEM., 46, 2003
|
|
1OXB
 
 | |
3FCD
 
 | |
3EVZ
 
 | |
3U90
 
 | | apoferritin: complex with SDS | | Descriptor: | CADMIUM ION, DODECYL SULFATE, Ferritin light chain | | Authors: | Liu, R, Bu, W, Xi, J, Mortazavi, S.R, Cheung-Lau, J.C, Dmochowski, I.J, Loll, P.J. | | Deposit date: | 2011-10-17 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Beyond the detergent effect: a binding site for sodium dodecyl sulfate (SDS) in mammalian apoferritin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AVA
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3GPK
 
 | |
3D0C
 
 | | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution | | Descriptor: | Dihydrodipicolinate synthase | | Authors: | Satyanarayana, L, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-01 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution.
To be Published
|
|
3CBN
 
 | |
3HP0
 
 | |
3G1W
 
 | |
3H49
 
 | |
3HDP
 
 | |
