1HDU
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HEE
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with L-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, L-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HDQ
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with D-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1LAH
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
6L7Q
 
 | |
6IWB
 
 | | Crystal structure of a computationally designed protein (LD3) in complex with BCL-2 | | Descriptor: | Apolipoprotein E, Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, SULFATE ION | | Authors: | Kim, S, Kwak, M.J, Oh, B.-H, Correia, B.E, Gainza, P. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy.
Nat.Biotechnol., 38, 2020
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1DU3
 
 | | Crystal structure of TRAIL-SDR5 | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND, ZINC ION | | Authors: | Cha, S.-S, Sung, B.-J, Oh, B.-H. | | Deposit date: | 2000-01-14 | | Release date: | 2000-09-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity
J.Biol.Chem., 275, 2000
|
|
1E3V
 
 | | Crystal structure of ketosteroid isomerase from Psedomonas putida complexed with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Kim, J.-S, Oh, B.-H. | | Deposit date: | 2000-06-24 | | Release date: | 2001-03-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1O9P
 
 | |
1O9Q
 
 | |
1OHT
 
 | | Peptidoglycan recognition protein LB | | Descriptor: | 1,2-ETHANEDIOL, CG14704 PROTEIN, L(+)-TARTARIC ACID, ... | | Authors: | Kim, M.-S, Byun, M, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Peptidoglycan Recognition Protein Lb from Drosophila Melanogaster
Nat.Immunol., 4, 2003
|
|
1OGF
 
 | | The Structure of Bacillus subtilis RbsD complexed with glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OHU
 
 | | Structure of Caenorhabditis elegans CED-9 | | Descriptor: | APOPTOSIS REGULATOR CED-9 | | Authors: | Jeong, J.-S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Unique Structural Features of a Bcl-2 Family Protein Ced-9 and Biophysical Characterization of Ced-9/Egl-1 Interactions
Cell Death Differ., 10, 2003
|
|
1OGE
 
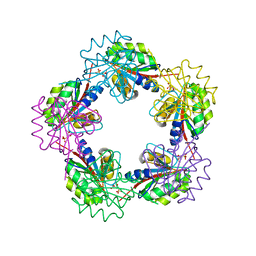 | | The Structure of Bacillus subtilis RbsD complexed with Ribose 5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OGC
 
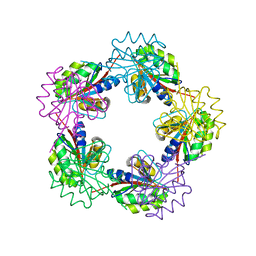 | | The Structure of Bacillus subtilis RbsD complexed with D-ribose | | Descriptor: | CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1OGD
 
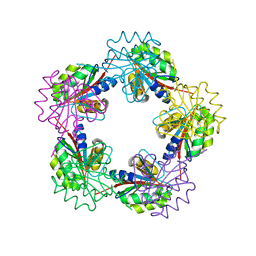 | | The Structure of Bacillus subtilis RbsD complexed with D-ribose | | Descriptor: | CHLORIDE ION, HIGH AFFINITY RIBOSE TRANSPORT PROTEIN RBSD, beta-D-ribopyranose | | Authors: | Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-04-30 | | Release date: | 2003-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Rbsd Leading to the Identification of Cytoplasmic Sugar-Binding Proteins with a Novel Folding Architecture
J.Biol.Chem., 278, 2003
|
|
1CVM
 
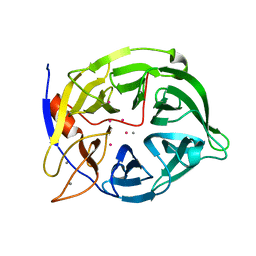 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1OH0
 
 | | CRYSTAL STRUCTURE OF KETOSTEROID ISOMERASE COMPLEXED WITH EQUILENIN | | Descriptor: | BETA-MERCAPTOETHANOL, EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Kim, K.-H, Cha, S.-S, Byun, M, Oh, B.-H. | | Deposit date: | 2003-05-21 | | Release date: | 2003-06-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Crystal Structures of Delta5-3-Ketosteroid Isomerase with and without a Reaction Intermediate Analogue
Biochemistry, 36, 1997
|
|
1OGX
 
 | | High Resolution Crystal Structure Of Ketosteroid Isomerase Mutant D40N(D38N, Ti Numbering) from Pseudomonas putida Complexed With Equilenin At 2.0 A Resolution. | | Descriptor: | EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-05-17 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbation of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1OCL
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COMPLEXED WITH MALONATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, MALONIC ACID | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OBL
 
 | |
1OBJ
 
 | |
1OCK
 
 | |
