4Y3Y
 
 | | Endothiapepsin in complex with fragment B42 | | Descriptor: | 2-AMINO-4-METHYL-PENTAN-1-OL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4Y3J
 
 | | Endothiapepsin in complex with fragment B30 | | Descriptor: | 4-METHYL-HISTIDINE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7MF0
 
 | | Co-crystal structure of PERK with inhibitor (R)-2-amino-N-cyclopropyl-5-(4-(2-(3,5-difluorophenyl)-2-hydroxyacetamido)-2-methylphenyl)nicotinamide | | Descriptor: | 2-amino-N-cyclopropyl-5-(4-{[(2R)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino}-2-methylphenyl)pyridine-3-carboxamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wiens, B, Koszelak-Rosenblum, M, Surman, M.D, Zhu, G, Mulvihill, M.J. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors.
Bioorg.Med.Chem.Lett., 43, 2021
|
|
7QVF
 
 | |
5JGA
 
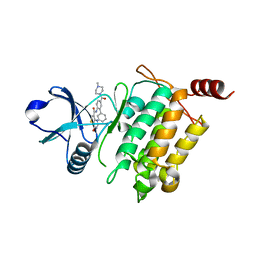 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 11c | | Descriptor: | N-[5-(4-methylpiperazine-1-carbonyl)[1,1'-biphenyl]-2-yl]-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
7QVC
 
 | |
7QWG
 
 | |
6NU4
 
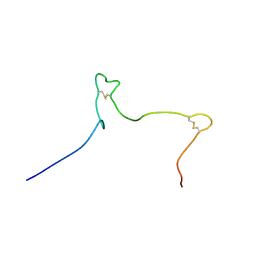 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
4ARA
 
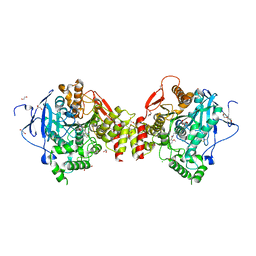 | | Mus musculus Acetylcholinesterase in complex with (R)-C5685 at 2.5 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6O0T
 
 | | Crystal structure of selenomethionine labelled tandem SAM domains (L446M:L505M:L523M mutant) from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O1B
 
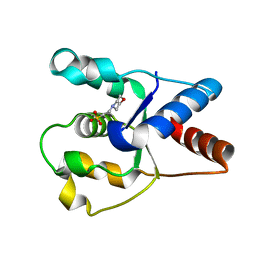 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
4GW2
 
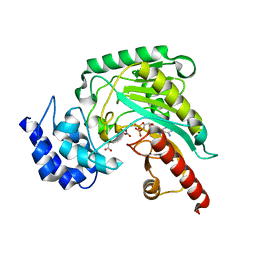 | | Crystal structure of arginine kinase in complex with L-ornithine, MgADP, and nitrate. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, L-ornithine, ... | | Authors: | Clark, S.A, Davulcu, O, Chapman, M.S. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Crystal structures of arginine kinase in complex with ADP, nitrate, and various phosphagen analogs.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
8H4V
 
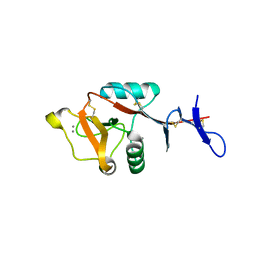 | | Mincle CRD complex with PGL trisaccharide | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol-(1-4)-6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-3-O-methyl-alpha-L-rhamnopyranose, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
8HB5
 
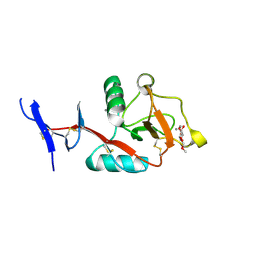 | | Crystal structure of Mincle in complex with HD-275 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-27 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
4GW0
 
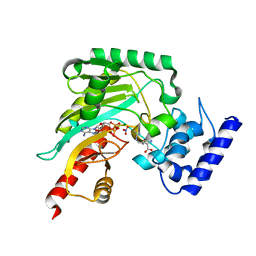 | | Crystal structure of arginine kinase in complex with imino-L-ornithine, MgADP, and nitrate. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, MAGNESIUM ION, ... | | Authors: | Clark, S.A, Davulcu, O, Chapman, M.S. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Crystal structures of arginine kinase in complex with ADP, nitrate, and various phosphagen analogs.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5JGB
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 10 | | Descriptor: | N-(2-methoxy-4-{[3-(4-methylpiperazin-1-yl)propyl]carbamoyl}phenyl)-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
6BX0
 
 | |
4GQI
 
 | | Synthesis of novel MT3 receptor ligands via unusual Knoevenagel condensation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-{(3R)-3-[2-(acetylamino)ethyl]-2-oxo-2,3-dihydro-1H-indol-5-yl}acetamide, ... | | Authors: | Volkova, M.S, Jensen, K.C, Lozinskaya, N.A, Sosonyuk, S.E, Proskurnina, M.V, Mesecar, A.D, Zefirov, N.S. | | Deposit date: | 2012-08-23 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Synthesis of novel МТ3 receptor ligands via an unusual Knoevenagel condensation.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4Y48
 
 | | Endothiapepsin in complex with fragment B50 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6BX1
 
 | |
5EBI
 
 | | Crystal structure of a DNA-RNA chimera in complex with Ba2+ ions: a case of unusual multi-domain twinning | | Descriptor: | BARIUM ION, DNA/RNA (5'-D(*C)-R(P*G)-D(P*C)-R(P*G)-D(P*C)-R(P*G)-3') | | Authors: | Gilski, M, Drozdzal, P, Kierzek, R, Jaskolski, M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic resolution structure of a chimeric DNA-RNA Z-type duplex in complex with Ba(2+) ions: a case of complicated multi-domain twinning.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7V2Y
 
 | | cryo-EM structure of yeast THO complex with Sub2 | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
7V2W
 
 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
