4OBD
 
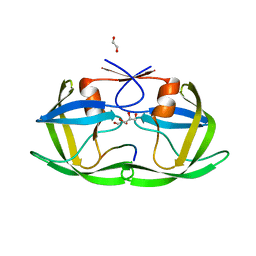 | |
4OQY
 
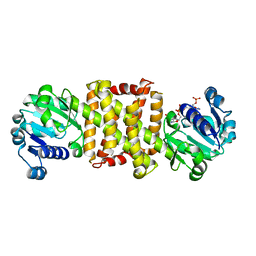 | | Streptomyces sp. GF3546 imine reductase | | Descriptor: | (S)-imine reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schneider, L.K, Huber, T, Gerhardt, S, Muller, M, Einsle, O. | | Deposit date: | 2014-02-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Direct Reductive Amination of Ketones: Structure and Activity of S-Selective Imine Reductases from Streptomyces.
CHEMCATCHEM, 2014
|
|
6B6C
 
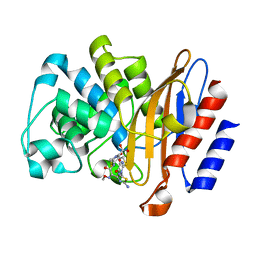 | |
4OU6
 
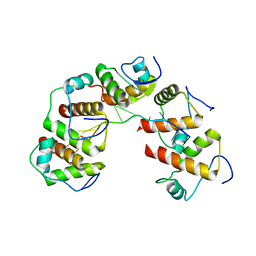 | | Crystal structure of DnaT84-153-dT10 ssDNA complex form 1 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | Authors: | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
6B6E
 
 | | Beta-Lactamase, mixed with Ceftriaxone, needles crystal form, 500ms | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
4OWZ
 
 | |
4OX8
 
 | |
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OW6
 
 | | Crystal structure of Diphtheria Toxin at acidic pH | | Descriptor: | Diphtheria toxin | | Authors: | Leka, O, Vallese, F, Pirazzini, M, Berto, P, Montecucco, C, Zanotti, G. | | Deposit date: | 2014-01-31 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diphtheria toxin conformational switching at acidic pH.
Febs J., 281, 2014
|
|
4O6U
 
 | | 0.89A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O9T
 
 | | Mechanism of transhydrogenase coupling proton translocation and hydride transfer | | Descriptor: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | Authors: | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | Deposit date: | 2014-01-02 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.079 Å) | | Cite: | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4OBK
 
 | |
3OT7
 
 | | Escherichia coli apo-manganese superoxide dismutase | | Descriptor: | Superoxide dismutase [Mn] | | Authors: | Whittaker, M.M, Lerch, T.F, Kirillova, O, Chapman, M.S, Whittaker, J.W. | | Deposit date: | 2010-09-10 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Subunit dissociation and metal binding by Escherichia coli apo-manganese superoxide dismutase.
Arch.Biochem.Biophys., 505, 2011
|
|
4OEN
 
 | | Crystal structure of the second substrate binding domain of a putative amino acid ABC transporter from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | ACETATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the second substrate binding domain of a putative amino acid ABC transporter from Streptococcus pneumoniae Canada MDR_19A
To be Published
|
|
4OFQ
 
 | | Structure of the C-terminal domain of the Streptococcus pyogenes antigen I/II-family protein AspA | | Descriptor: | CALCIUM ION, Putative cell surface protein | | Authors: | Hall, M, Nylander, A, Jenkinson, H.F, Persson, K. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of AspA (antigen I/II-family) protein from Streptococcus pyogenes.
FEBS Open Bio, 4, 2014
|
|
4OI3
 
 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
3SIJ
 
 | |
6B68
 
 | | Beta-Lactamase, 100ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
1QNY
 
 | | X-ray refinement of D2O soaked crystal of concanavalin A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb(Gilboa), A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct Determination of the Positions of Deuterium Atoms of Bound Water in Concanavalin a by Neutron Laue Crystallography
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4OP4
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea in the Zn bound form | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
4OUU
 
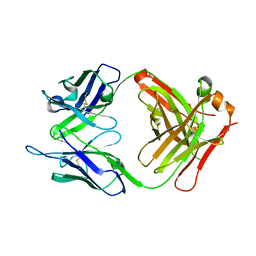 | | anti-MT1-MMP monoclonal antibody | | Descriptor: | anti_MT1-MMP Heavy chain, anti_MT1-MMP light chain | | Authors: | Udi, Y, Grossman, M, Solomonov, I, Dym, O, Rozenberg, H, Koziol, A, Cuniasse, P, Dive, V, Arroyo, A.G, Irit, S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2014-02-19 | | Release date: | 2014-12-17 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
4ORZ
 
 | |
3RUB
 
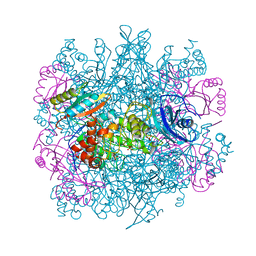 | | CRYSTAL STRUCTURE OF THE UNACTIVATED FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM TOBACCO REFINED AT 2.0-ANGSTROMS RESOLUTION | | Descriptor: | ASPARAGINE, RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, FORM III, ... | | Authors: | Schreuder, H, Cascio, D, Curmi, P.M.G, Chapman, M.S, Suh, S.W, Eisenberg, D.S. | | Deposit date: | 1990-05-25 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution.
J.Biol.Chem., 267, 1992
|
|
4OBZ
 
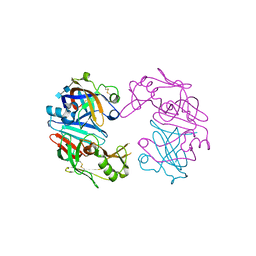 | | Structure of Cathepsin D with inhibitor 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide | | Descriptor: | 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OU7
 
 | | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | Authors: | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
