3EPN
 
 | | Crystal structure of Caulobacter crescentus ThiC complexed with imidazole ribonucleotide | | Descriptor: | 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, Thiamine biosynthesis protein thiC | | Authors: | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
3ERI
 
 | | First structural evidence of substrate specificity in mammalian peroxidases: Crystal structures of substrate complexes with lactoperoxidases from two different species | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sheikh, I.A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-10-02 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
3ETE
 
 | | Crystal structure of bovine glutamate dehydrogenase complexed with hexachlorophene | | Descriptor: | 2,2'-methanediylbis(3,4,6-trichlorophenol), GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, M, Smith, T.J. | | Deposit date: | 2008-10-07 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel Inhibitors Complexed with Glutamate Dehydrogenase: ALLOSTERIC REGULATION BY CONTROL OF PROTEIN DYNAMICS
J.Biol.Chem., 284, 2009
|
|
3ERH
 
 | | First structural evidence of substrate specificity in mammalian peroxidases: Crystal structures of substrate complexes with lactoperoxidases from two different species | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Sheikh, I.A, Singh, N, Singh, A.K, Sinha, M, Singh, S.B, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-10-02 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
3E2V
 
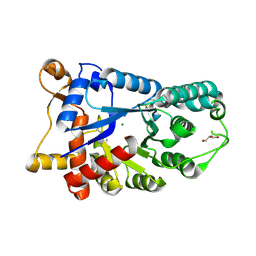 | | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae | | Descriptor: | 3'-5'-exonuclease, GLYCEROL, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae
To be Published
|
|
7YDS
 
 | | The structure of the bispecific antibody targeted PD-L1 and 4-1BB | | Descriptor: | Anti-PDL1-VH-CH1, Anti-PDL1-VL-CL, Programmed cell death 1 ligand 1 | | Authors: | Gao, Y, Zhu, M, Liu, W.T, Cheng, L.S, Zhu, Z.L, Niu, L.W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A bispecific antibody targeted PD-L1 and 4-1BB induces a potent antitumor immune activity in colorectal cancer without systemic toxicity
To Be Published
|
|
7YHN
 
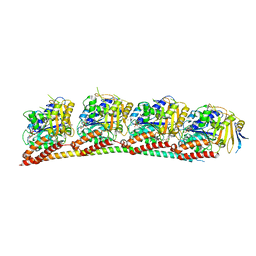 | | ANTI-TUMOR AGENT Y48 IN COMPLEX WITH TUBULIN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methyl-3-[(4-methylphenyl)sulfonylamino]-~{N}-[(6-methylpyridin-3-yl)methyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Du, T, Ji, M, Hou, Z, Lin, S, Zhang, J, Wu, D, Zhang, K, Lu, D, Xu, H, Chen, X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Benzamide Derivatives as Potent and Orally Active Tubulin Inhibitors Targeting the Colchicine Binding Site.
J.Med.Chem., 65, 2022
|
|
3E7V
 
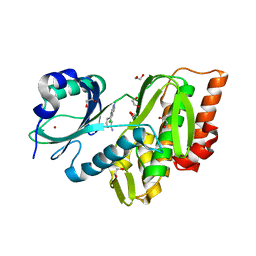 | | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-aminophenyl)-N-(3-chlorophenyl)pyrazolo[1,5-a]pyrimidin-5-amine, NICKEL (II) ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand
To be Published
|
|
3E80
 
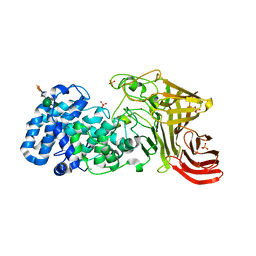 | |
3EBJ
 
 | | Crystal structure of an avian influenza virus protein | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein | | Authors: | Yuan, P, Bartlam, M, Lou, Z, Chen, S, Rao, Z, Liu, Y. | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site
Nature, 458, 2009
|
|
3EEI
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Ho, M, Rinaldo-matthis, A, Brown, R.L, Norris, G.E, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-09-04 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A
To be Published
|
|
3EFB
 
 | | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri | | Descriptor: | ACETIC ACID, Probable sor-operon regulator | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri
To be Published
|
|
3EL5
 
 | |
3EM4
 
 | | Crystal structure of atazanavir (ATV) in complex with I50L/A71V drug-resistant HIV-1 protease | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N, Royer, C, Schiffer, C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Structural studies on atazanavir-specific I50L drug-resistant HIV-1 protease mutant
To be Published
|
|
3EOK
 
 | |
3DD0
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-06-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
3DFS
 
 | |
7UQA
 
 | | Crystal structure of the small Ultra-Red Fluorescent Protein (smURFP) | | Descriptor: | CHLORIDE ION, SODIUM ION, small Ultra-Red Fluorescent Protein (smURFP) | | Authors: | Maiti, A, Buffalo, C.Z, Saurabh, S, Montecinos-Franjola, F, Hachey, J.S, Conlon, W.J, Tran, G.N, Drobizhev, M, Moerner, W.E, Ghosh, P, Matsuo, H, Tsien, R.Y, Lin, J.Y, Rodriguez, E.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural and photophysical characterization of the small ultra-red fluorescent protein.
Nat Commun, 14, 2023
|
|
3DHZ
 
 | |
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3DHN
 
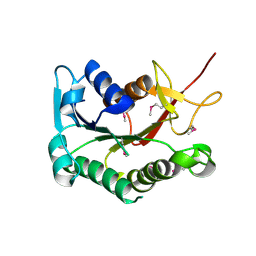 | | Crystal structure of the putative epimerase Q89Z24_BACTN from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR310. | | Descriptor: | NAD-dependent epimerase/dehydratase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Wang, D, Ciccosanti, C, Foote, L.E, Janjua, H, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative epimerase Q89Z24_BACTN from Bacteroides thetaiotaomicron.
To be Published
|
|
3DHT
 
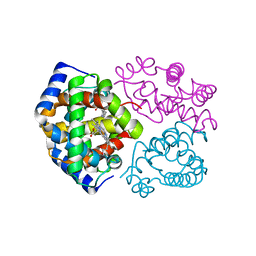 | | The Crystal Structure Determination of Rat (rattus norvegicus) Hemoglobin | | Descriptor: | Hemoglobin subunit alpha-1/2, Hemoglobin subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Neelagandan, K, Sathya Moorthy, P, Balasubramanian, M, Sundaresan, S, Ponnuswamy, M.N. | | Deposit date: | 2008-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The Crystal Structure Determination of Rat (rattus norvegicus) Hemoglobin
To be Published
|
|
3DMI
 
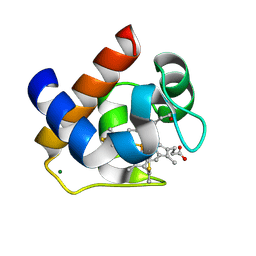 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
3D9S
 
 | | Human Aquaporin 5 (AQP5) - High Resolution X-ray Structure | | Descriptor: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | Authors: | Horsefield, R, Norden, K, Fellert, M, Backmark, A, Tornroth-Horsefield, S, Terwisscha Van Scheltinga, A.C, Kvassman, J, Kjellbom, P, Johanson, U, Neutze, R. | | Deposit date: | 2008-05-27 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution x-ray structure of human aquaporin 5
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
