4O2F
 
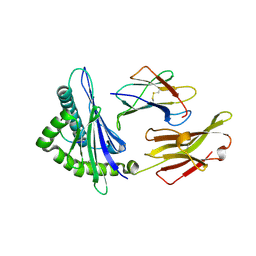 | | A peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
4O4J
 
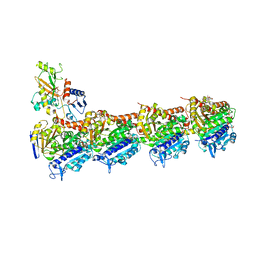 | | Tubulin-Peloruside A complex | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside A.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4O5W
 
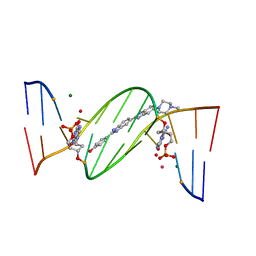 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5Z
 
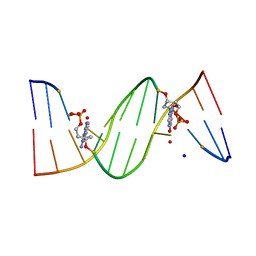 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), SODIUM ION | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O3C
 
 | | Crystal structure of the GLUA2 ligand-binding domain in complex with L-aspartate at 1.50 A resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, K, Kaern, A.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O4H
 
 | | Tubulin-Laulimalide complex | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside a.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4O3T
 
 | | Zymogen HGF-beta/MET with Zymogen Activator Peptide ZAP.14 | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor, PENTAETHYLENE GLYCOL, ... | | Authors: | Eigenbrot, C, Landgraf, K.E, Steffek, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | An allosteric switch for pro-HGF/Met signaling using zymogen activator peptides.
Nat.Chem.Biol., 10, 2014
|
|
4O4L
 
 | | Tubulin-Peloruside A-Epothilone A complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, EPOTHILONE A, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside a.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4O3U
 
 | |
3UXM
 
 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 5'-deoxy-5'-fluorothymidine, MAGNESIUM ION, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
4O4O
 
 | | Crystal structure of phycobiliprotein lyase CpcT | | Descriptor: | MAGNESIUM ION, Phycocyanobilin lyase CpcT | | Authors: | Zhou, W, Ding, W.-L, Zeng, X.-l, Dong, L.-L, Zhao, B, Zhou, M, Scheer, H, Zhao, K.-H, Yang, X. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of the Phycobiliprotein Lyase CpcT.
J.Biol.Chem., 289, 2014
|
|
2XYO
 
 | | Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
4O4Q
 
 | | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution
To be Published
|
|
4O55
 
 | |
4O5A
 
 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
4O8E
 
 | | Crystal structure of the complex of type I ribosome inactivating protein from Momordica balsamina with uridine triphosphate at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Pandey, S, Yamini, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-27 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein from Momordica balsamina with uridine triphosphate at 2.0 A resolution
To be Published
|
|
4O79
 
 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 10 minutes | | Descriptor: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O90
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.6A resolution | | Descriptor: | Chorismate synthase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Chaudhary, A, Singh, N, Shukla, P.K, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-31 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.6A resolution
To be Published
|
|
2YNT
 
 | | GIM-1-3Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN, GLYCEROL, ZINC ION | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
4OHN
 
 | | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, HISTIDINE | | Authors: | Brunzelle, J.S, Wawrzak, W, Yim, Y, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine
To be Published
|
|
2YNW
 
 | | GIM-1-2Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN, GLYCEROL, SULFATE ION, ... | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
1EKJ
 
 | |
4QFZ
 
 | | Crystal structure of the tetrameric dGTP/dTTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
4QHI
 
 | | Crystal structure of Methanocaldococcus jannaschii selecase mutant R36W | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1RFG
 
 | | Crystal Structure of Human Purine Nucleoside Phosphorylase Complexed with Guanosine | | Descriptor: | GUANOSINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Canduri, F, Silva, R.G, Dos Santos, D.M, Palma, M.S, Basso, L.A, Santos, D.S, de Azevedo, W.F. | | Deposit date: | 2003-11-09 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human PNP complexed with ligands.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
