3DRU
 
 | | Crystal Structure of Gly117Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
3DP5
 
 | | Crystal structure of Geobacter sulfurreducens OmcF with N-terminal Strep-tag II | | Descriptor: | Cytochrome c family protein, HEME C, SULFATE ION | | Authors: | Lukat, P, Hoffmann, M, Einsle, O. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-02 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal packing of the c(6)-type cytochrome OmcF from Geobacter sulfurreducens is mediated by an N-terminal Strep-tag II
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
3DU3
 
 | | E(L212)A, D(L213)A, A(M249)Y triple mutant structure of photosynthetic reaction center | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2008-07-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | E(L212)A, D(L213)A, A(M249)Y triple mutant structure of photosynthetic reaction center
To be Published
|
|
3DY5
 
 | | Allene oxide synthase 8R-lipoxygenase from Plexaura homomalla | | Descriptor: | Allene oxide synthase-lipoxygenase protein, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gilbert, N.C, Niebuhr, M, Tsuruta, H, Newcomer, M.E. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | A covalent linker allows for membrane targeting of an oxylipin biosynthetic complex.
Biochemistry, 47, 2008
|
|
3E07
 
 | | Crystal structure of spatzle cystine knot | | Descriptor: | GLYCEROL, Protein spaetzle | | Authors: | Hoffmann, A, Funkner, A, Neumann, P, Juhnke, S, Walther, M, Schierhorn, A, Weininger, U, Balbach, J, Reuter, G, Stubbs, M.T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biophysical Characterization of Refolded Drosophila Spatzle, a Cystine Knot Protein, Reveals Distinct Properties of Three Isoforms
J.Biol.Chem., 283, 2008
|
|
3DT5
 
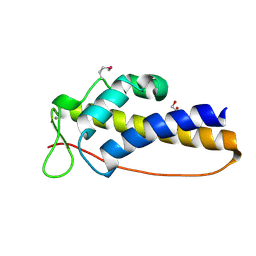 | | C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Uncharacterized protein AF_0924 | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-14 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray crystal structure of C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus.
To be Published
|
|
3DFT
 
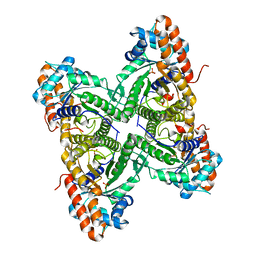 | |
3DL9
 
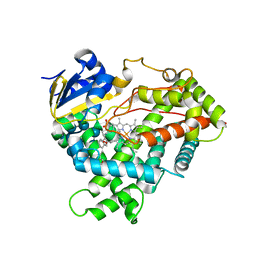 | | Crystal structure of CYP2R1 in complex with 1-alpha-hydroxy-vitamin D2 | | Descriptor: | (1S,3R,5Z,7E,22E)-9,10-secoergosta-5,7,10,22-tetraene-1,3-diol, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Cytochrome P450 2R1, ... | | Authors: | Strushkevich, N.V, Tempel, W, Gilep, A.A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Wilkstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.721 Å) | | Cite: | Crystal structure of CYP2R1 in complex with 1-alpha-hydroxy-vitamin D2.
To be Published
|
|
3DSY
 
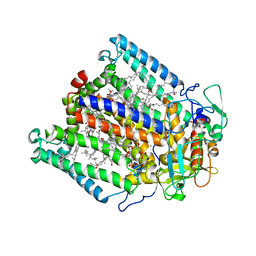 | |
3DVF
 
 | |
3DVL
 
 | |
3DXF
 
 | | Crystal structure of the HSCARG R37A mutant | | Descriptor: | NmrA-like family domain-containing protein 1 | | Authors: | Li, Y, Meng, G, Dai, X, Luo, M, Zheng, X. | | Deposit date: | 2008-07-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NADPH is an allosteric regulator of HSCARG
J.Mol.Biol., 387, 2009
|
|
3DY4
 
 | | Crystal structure of yeast 20S proteasome in complex with spirolactacystin | | Descriptor: | Omuralide, open form, Proteasome component C1, ... | | Authors: | Groll, M, Balskus, E, Jacobsen, E. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of spiro beta-lactone proteasome inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
3EGC
 
 | | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis | | Descriptor: | putative ribose operon repressor | | Authors: | Bonanno, J.B, Patskovsky, Y, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative ribose operon repressor from Burkholderia thailandensis
To be Published
|
|
3ED8
 
 | |
3EKC
 
 | |
3EL9
 
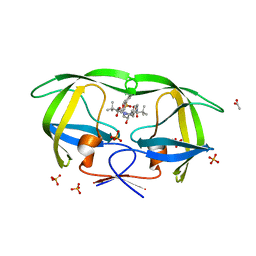 | | Crystal structure of atazanavir (ATV) in complex with a multidrug HIV-1 protease (V82T/I84V) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N, Schiffer, C. | | Deposit date: | 2008-09-21 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3EF7
 
 | |
3ELK
 
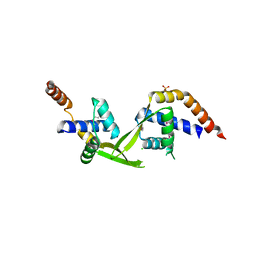 | | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator TA0346 | | Authors: | Grantz Saskova, K, Chruszcz, M, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-22 | | Release date: | 2008-09-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum
To be Published
|
|
3EIP
 
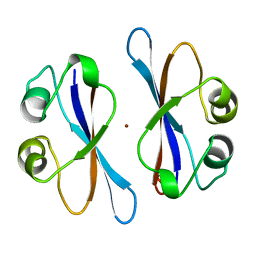 | | CRYSTAL STRUCTURE OF COLICIN E3 IMMUNITY PROTEIN: AN INHIBITOR TO A RIBOSOME-INACTIVATING RNASE | | Descriptor: | PROTEIN (COLICIN E3 IMMUNITY PROTEIN), ZINC ION | | Authors: | Li, C, Zhao, D, Djebli, A, Shoham, M. | | Deposit date: | 1999-03-29 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase.
Structure Fold.Des., 7, 1999
|
|
3H5E
 
 | |
3EOZ
 
 | | Crystal Structure of Phosphoglycerate Mutase from Plasmodium Falciparum, PFD0660w | | Descriptor: | GLYCEROL, PHOSPHATE ION, putative Phosphoglycerate mutase | | Authors: | Wernimont, A.K, Tempel, W, Lam, A, Zhao, Y, Lew, J, Lin, Y.H, Wasney, G, Vedadi, M, Kozieradzki, I, Cossar, D, Schapira, M, Weigelt, J, Arrowsmith, C.H, Bochkarev, A, Edwards, A.M, Hui, R, Pizarro, J, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a new phosphatase from Plasmodium.
Mol.Biochem.Parasitol., 179, 2011
|
|
3EPR
 
 | | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae. | | Descriptor: | GLYCEROL, Hydrolase, haloacid dehalogenase-like family, ... | | Authors: | Ramagopal, U.A, Toro, R, Dickey, M, Tang, B.K, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae.
To be Published
|
|
3EPW
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase in complex with the inhibitor (2R,3R,4S)-1-[(4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-2-(hydroxymethyl)pyrrolidin-3,4-diol | | Descriptor: | 7-(((2R,3R,4S)-3,4-dihydroxy-2-(hydroxymethyl)pyrrolidin-1-yl)methyl)-3H-pyrrolo[3,2-d]pyrimidin-4(5H)-one, CALCIUM ION, IAG-nucleoside hydrolase, ... | | Authors: | Versees, W, Goeminne, A, Berg, M, Vandemeulebroucke, A, Haemers, A, Augustyns, K, Steyaert, J. | | Deposit date: | 2008-09-30 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of T. vivax nucleoside hydrolase in complex with new potent and specific inhibitors.
Biochim.Biophys.Acta, 1794, 2009
|
|
3EQE
 
 | | Crystal structure of the YubC protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR112. | | Descriptor: | FE (III) ION, Putative cystein dioxygenase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Benach, J, Forouhar, F, Clayton, G, Cooper, B, Wang, H, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the YubC protein from Bacillus subtilis.
To be Published
|
|
