7Y06
 
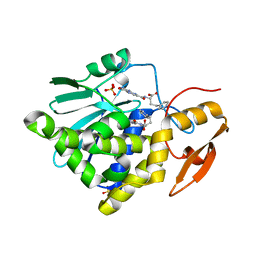 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-4-phenylbutanoic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-4-phenyl-butanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y03
 
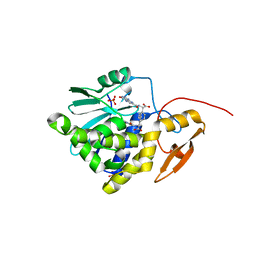 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-2-phenylacetic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-2-phenyl-ethanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y07
 
 | | Crystal structure of Ricin A chain bound with (S)-2-amino-N-(1-hydroxy-3-phenylpropan-2-yl)-4-oxo-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-azanyl-4-oxidanylidene-N-[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]-3H-pteridine-7-carboxamide, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y08
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanine | | Descriptor: | N-[(2-amino-4-oxo-1,4-dihydropteridin-7-yl)carbonyl]glycyl-L-phenylalanine, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
3B1X
 
 | | Crystal structure of an S. thermophilus NFeoB E66A mutant bound to GMPPNP | | Descriptor: | Ferrous iron uptake transporter protein B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3B4M
 
 | | Crystal Structure of Human PABPN1 RRM | | Descriptor: | Polyadenylate-binding protein 2 | | Authors: | Ge, H, Zhou, D, Teng, M, Niu, L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure and possible dimerization of the single RRM of human PABPN1
Proteins, 71, 2008
|
|
3B0P
 
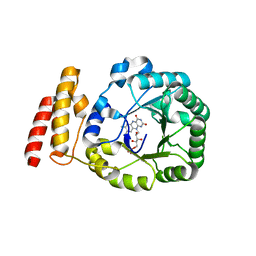 | | tRNA-dihydrouridine synthase from Thermus thermophilus | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-12 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B76
 
 | | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase LNX, SODIUM ION | | Authors: | Ugochukwu, E, Burgess-Brown, N, Berridge, G, Elkins, J, Bunkoczi, G, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Gileadi, O, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor.
To be Published
|
|
7XSG
 
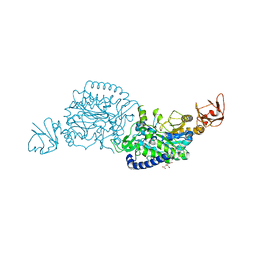 | | Crystal structure of ClAgl29B | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7YIM
 
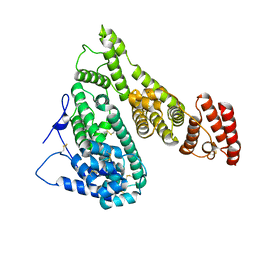 | | Cryo-EM structure of human Alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein | | Authors: | Liu, N, Liu, K, Wu, C, Liu, Z, Li, M, Wang, J, Wang, H.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
7XSF
 
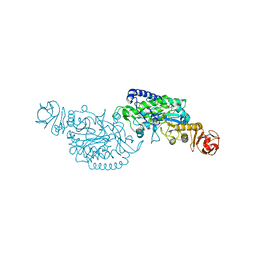 | | Crystal structure of ClAgl29A | | Descriptor: | Alpha-L-fucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7XSH
 
 | | Crystal structure of ClAgl29B bound with L-glucose | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
3B6P
 
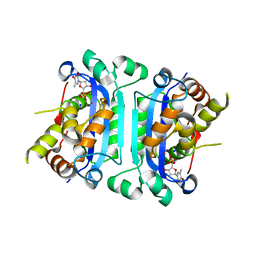 | | Structure of TREX1 in complex with a nucleotide and inhibitor ions (sodium and zinc) | | Descriptor: | SODIUM ION, THYMIDINE-5'-PHOSPHATE, Three prime repair exonuclease 1, ... | | Authors: | Brucet, M, Querol-Audi, J, Fita, I, Celada, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical studies of TREX1 inhibition by metals. Identification of a new active histidine conserved in DEDDh exonucleases.
Protein Sci., 17, 2008
|
|
7Y0Z
 
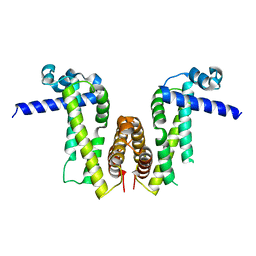 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
3BBR
 
 | |
7VB6
 
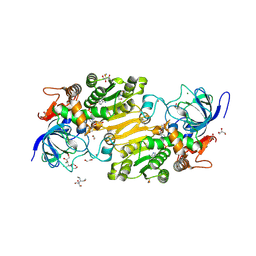 | | Crystal structure of hydroxynitrile lyase from Linum usitatissium complexed with (R)-2-hydroxy-2-methylbutanenitrile | | Descriptor: | (2R)-2-methyl-2-oxidanyl-butanenitrile, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
3BA7
 
 | |
7VK2
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.0 A resolution -9 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Gul, M. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VB3
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aliphatic (R)-hydroxynitrile lyase, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
7VB5
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum complexed with acetone cyanohydrin | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXY-2-METHYLPROPANENITRILE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
3BC1
 
 | | Crystal Structure of the complex Rab27a-Slp2a | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-27A, ... | | Authors: | Chavas, L.M.G, Ihara, K, Kawasaki, M, Wakatsuki, S. | | Deposit date: | 2007-11-12 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Rab27 recruitment by its effectors: structure of Rab27a bound to Exophilin4/Slp2-a
Structure, 16, 2008
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
3BDU
 
 | | Crystal structure of protein Q6D8G1 at the resolution 1.9 A. Northeast Structural Genomics Consortium target EwR22A. | | Descriptor: | Putative lipoprotein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein Q6D8G1 at the resolution 1.9 A.
To be Published
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
