3C9E
 
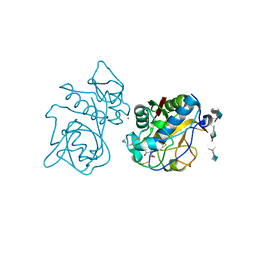 | | Crystal structure of the cathepsin K : chondroitin sulfate complex. | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Kienetz, M, Cherney, M.M, James, M.N.G, Bromme, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex.
J.Mol.Biol., 383, 2008
|
|
7ZBO
 
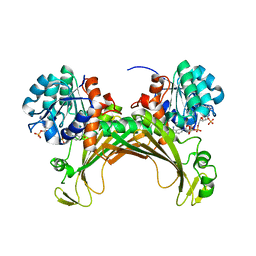 | | Amine Dehydrogenase MATOUAmDH2 in complex with NADP+ | | Descriptor: | Amine Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M, Ducrot, L, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.
Chembiochem, 23, 2022
|
|
7ZEG
 
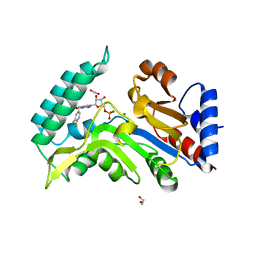 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7GMP | | Descriptor: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
7ZEE
 
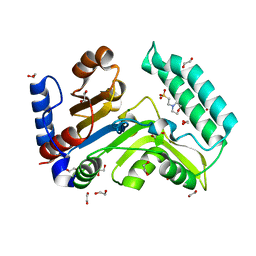 | | Human cytosolic 5' nucleotidase IIIB | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methylguanosine phosphate-specific 5'-nucleotidase, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
3BP1
 
 | | Crystal structure of putative 7-cyano-7-deazaguanine reductase QueF from Vibrio cholerae O1 biovar eltor | | Descriptor: | GUANINE, MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-08 | | Last modified: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of the nitrile reductase QueF combined with molecular simulations provide insight into enzyme mechanism.
J.Mol.Biol., 404, 2010
|
|
7Z51
 
 | | Tick-borne encephalitis virus Kuutsalo-14 | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Pulkkinen, L.I.A, Barrass, S.V, Anastasina, M, Butcher, S.J. | | Deposit date: | 2022-03-07 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular Organisation of Tick-Borne Encephalitis Virus.
Viruses, 14, 2022
|
|
3BRE
 
 | | Crystal Structure of P.aeruginosa PA3702 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Probable two-component response regulator | | Authors: | De, N, Pirruccello, M, Krasteva, P.V, Bae, N, Raghavan, R.V, Sondermann, H. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation-independent regulation of the diguanylate cyclase WspR.
Plos Biol., 6, 2008
|
|
3DKM
 
 | | Crystal structure of the HECTD1 CPH domain | | Descriptor: | E3 ubiquitin-protein ligase HECTD1 | | Authors: | Walker, J.R, Qiu, L, Li, Y, Bountra, C, Wolkstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the CPH domain of the E3 ubiquitin-protein ligase HECTD1.
To be Published
|
|
3DME
 
 | | Crystal structure of conserved exported protein from Bordetella pertussis. NorthEast Structural Genomics target BeR141 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L(+)-TARTARIC ACID, conserved exported protein | | Authors: | Seetharaman, J, Abashidze, M, Wang, D, Janjua, H, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of conserved exported protein from Bordetella pertussis. NorthEast Structural Genomics target BeR141
To be Published
|
|
3DA1
 
 | | X-Ray structure of the glycerol-3-phosphate dehydrogenase from Bacillus halodurans complexed with FAD. Northeast Structural Genomics Consortium target BhR167. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glycerol-3-phosphate dehydrogenase | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray structure of the glycerol-3-phosphate dehydrogenase from Bacillus halodurans complexed with FAD. Northeast Structural Genomics Consortium target BhR167.
To be Published
|
|
3DAX
 
 | | Crystal structure of human CYP7A1 | | Descriptor: | Cytochrome P450 7A1, PROTOPORPHYRIN IX CONTAINING FE, UNKNOWN ATOM OR ION | | Authors: | Strushkevich, N.V, Tempel, W, Dombrovski, L, Dong, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Wilkstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human CYP7A1
To be Published
|
|
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
3DBQ
 
 | | Crystal structure of TTK kinase domain | | Descriptor: | Dual specificity protein kinase TTK | | Authors: | Wang, W, Yang, Y.T, Gao, Y.F, Zhu, S.C, Wang, F, Old, W, Xu, Q.B, Resing, K, Ahn, N, Lei, M, Liu, X.D. | | Deposit date: | 2008-06-02 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into Mps1 Kinase Activation
J.CELL.MOL.MED., 13, 2008
|
|
3DC9
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-06-03 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
3DQG
 
 | | Peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans. | | Descriptor: | Heat shock 70 kDa protein F | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans.
To be Published
|
|
7YYH
 
 | | Structure of the human CCANdeltaT CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7YWX
 
 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7ZEH
 
 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7Guanine | | Descriptor: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, D-ribulose, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
3DFA
 
 | | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum | | Descriptor: | Calcium-dependent protein kinase cgd3_920 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Hassanali, A, Khuu, C, Alam, Z, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum.
To be Published
|
|
3DFO
 
 | |
3DB9
 
 | | Crystal structure of UPF0317 protein Atu3911 from Agrobacterium tumefaciens. NorthEast Strcutural Genomics target AtR186 | | Descriptor: | UPF0317 protein Atu3911 | | Authors: | Seetharaman, J, Abashidze, M, Wang, D, Janjua, H, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of UPF0317 protein Atu3911 from Agrobacterium tumefaciens. NorthEast Strcutural Genomics target AtR186 (CASP Target)
TO BE PUBLISHED
|
|
3DHR
 
 | | Crystal Structure Determination of Methemoglobin from Pigeon at 2 Angstrom Resolution (Columba livia) | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sathya Moorthy, P, Neelagandan, K, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2008-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Purification, Crystallization and X-ray Diffraction Studies of Pigeon Hemoglobin
To be Published
|
|
3DEG
 
 | | Complex of elongating Escherichia coli 70S ribosome and EF4(LepA)-GMPPNP | | Descriptor: | 30S RNA helix 14, 30S RNA helix 8, 30S ribosomal protein S12, ... | | Authors: | Connell, S.R, Topf, M, Qin, Y, Wilson, D.N, Mielke, T, Fucini, P, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation
Nat.Struct.Mol.Biol., 15, 2008
|
|
3DFQ
 
 | |
7Z52
 
 | | Human NEXT dimer - focused reconstruction of the single MTR4 | | Descriptor: | Exosome RNA helicase MTR4, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*UP*UP*UP*UP*U)-3'), ... | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
