4Q7M
 
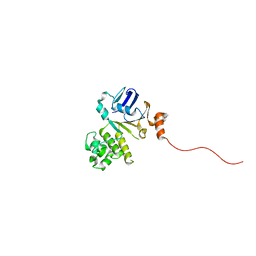 | | Structure of NBD288-Avi of TM287/288 | | Descriptor: | Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Bukowska, M.A, Hohl, M, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-25 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Transporter Motor Taken Apart: Flexibility in the Nucleotide Binding Domains of a Heterodimeric ABC Exporter.
Biochemistry, 54, 2015
|
|
4QC8
 
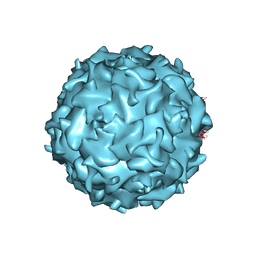 | | Structural annotation of pathogenic bovine Parvovirus-1 | | Descriptor: | VP2 | | Authors: | Kailasan, S, Halder, S, Gurda, B.L, Bladek, H, Chipman, P.R, McKenna, R, Brown, K, Agbandje-McKenna, M. | | Deposit date: | 2014-05-09 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an enteric pathogen, bovine parvovirus.
J.Virol., 89, 2015
|
|
4Q8S
 
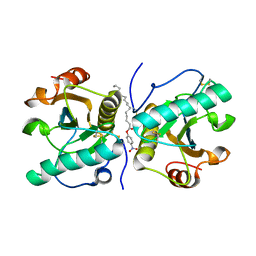 | | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrophenyl hexadecanoate, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution
To be Published
|
|
4Q9E
 
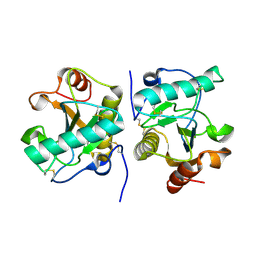 | | Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution
to be published
|
|
4QA9
 
 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | Authors: | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4QGE
 
 | | phosphodiesterase-9A in complex with inhibitor WYQ-C36D | | Descriptor: | MAGNESIUM ION, N~2~-(1-cyclopentyl-4-oxo-4,7-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)-N-(4-methoxyphenyl)-D-alaninamide, Phosphodiesterase 9A, ... | | Authors: | Shao, Y.-X, Huang, M, Cui, W, Ke, H. | | Deposit date: | 2014-05-22 | | Release date: | 2014-12-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Phosphodiesterase 9A Inhibitor as a Potential Hypoglycemic Agent.
J.Med.Chem., 57, 2014
|
|
4QGX
 
 | | Crystal structure of the R132K:R111L:L121E mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.47 angstrom resolution | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
1E7M
 
 | |
4QH5
 
 | | The GLIC pentameric Ligand-Gated Ion Channel (wild-type) crystallized in phosphate buffer | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Delarue, M, Sauguet, L. | | Deposit date: | 2014-05-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of potential modulation sites in the extracellular domain of the prokaryotic pentameric proton-gated ion channel GLIC
Acta Crystallogr.,Sect.D, 2015
|
|
4QFX
 
 | | Crystal structure of the tetrameric dGTP/dATP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
4QGV
 
 | | Crystal structure of the R132K:R111L mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.73 angstrom resolution. | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
4QG2
 
 | | Crystal structure of the tetrameric GTP/dATP/ATP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
4QNM
 
 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QHG
 
 | | Crystal structure of Methanocaldococcus jannaschii dimeric selecase | | Descriptor: | GLYCEROL, Uncharacterized protein MJ1213, ZINC ION | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4QO3
 
 | |
4QO8
 
 | | Lactate Dehydrogenase A in complex with substituted 3-Hydroxy-2-mercaptocyclohex-2-enone compound 104 | | Descriptor: | (5S)-2-[(2-chlorophenyl)sulfanyl]-5-(2,6-dichlorophenyl)-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Identification of substituted 3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4QI9
 
 | | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate
To be Published
|
|
4QAJ
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4QOD
 
 | | The value crystal structure of apo quinone reductase 2 at 1.35A | | Descriptor: | GLYCEROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The value crystal structure of apo quinone reductase 2 at 1.35A
To be Published
|
|
1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
4QG0
 
 | | Crystal structure of the tetrameric dGTP/dUTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DEOXYURIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
1B5G
 
 | | HUMAN THROMBIN COMPLEXED WITH NOVEL SYNTHETIC PEPTIDE MIMETIC INHIBITOR AND HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, SODIUM ION, ... | | Authors: | St Charles, R, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
3TRQ
 
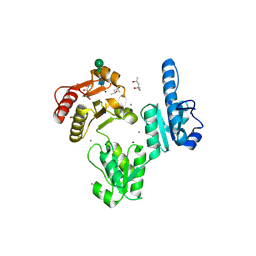 | | Crystal structure of native rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Glycosylation of Skeletal Calsequestrin: IMPLICATIONS FOR ITS FUNCTION.
J.Biol.Chem., 287, 2012
|
|
4PQ8
 
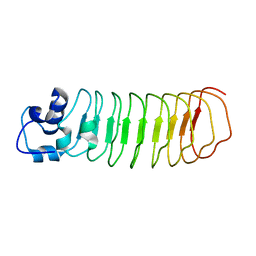 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR465 | | Descriptor: | CHLORIDE ION, DESIGNED PROTEIN OR465 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal Structure of Engineered Protein OR465.
To be Published
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
