4P1G
 
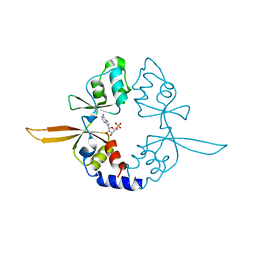 | | Crystal structure of the Bateman domain of murine magnesium transporter CNNM2 bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Abascal-Palacios, G, Diercks, T, Oyenarte, I, Ereno-Orbea, J, Encinar, J.A, Spiwok, V, Terashima, H, Accardi, A, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To Be Published
|
|
4P4G
 
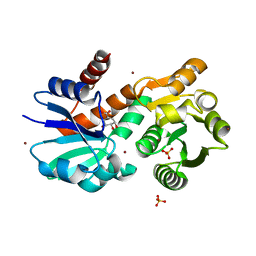 | | Crystal Structure of Mycobacterium tuberculosis Shikimate Dehydrogenase | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, BROMIDE ION, SULFATE ION, ... | | Authors: | Lalgondar, M, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-03-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Dehydrogenase
To Be Published
|
|
4P6Q
 
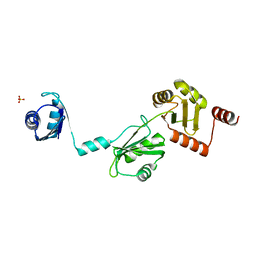 | | The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA Recognition Motifs | | Descriptor: | Msx2-interacting protein, SULFATE ION | | Authors: | Arieti, F, Gabus, C, Tambalo, M, Huet, T, Round, A, Thore, S. | | Deposit date: | 2014-03-25 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs.
Nucleic Acids Res., 42, 2014
|
|
4P6Y
 
 | |
4PG3
 
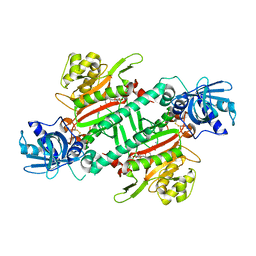 | | Crystal structure of KRS complexed with inhibitor | | Descriptor: | LYSINE, Lysine--tRNA ligase, cladosporin | | Authors: | Sharma, A, Yogavel, M, Khan, S, Sharma, A, Belrhali, H. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.
J. Struct. Funct. Genomics, 15, 2014
|
|
4PB6
 
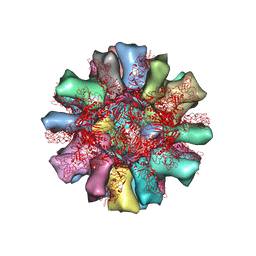 | |
2WP6
 
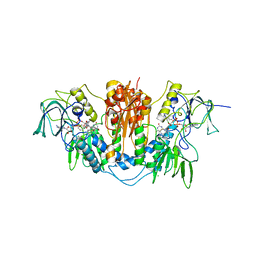 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00071494) | | Descriptor: | (4S)-3-BENZYL-6-CHLORO-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
4PF4
 
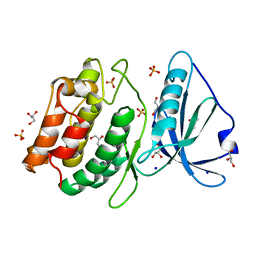 | | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277 | | Descriptor: | Death-associated protein kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Temmerman, K, Simon, B, Wilmanns, M. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277
To Be Published
|
|
3PTJ
 
 | | Structural and functional Analysis of Arabidopsis thaliana thylakoid lumen protein AtTLP18.3 | | Descriptor: | UPF0603 protein At1g54780, chloroplastic | | Authors: | Wu, H.Y, Liu, M.S, Lin, T.P, Cheng, Y.S. | | Deposit date: | 2010-12-03 | | Release date: | 2011-10-26 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen
Plant Physiol., 157, 2011
|
|
4P3S
 
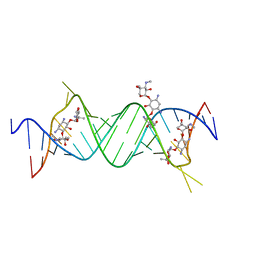 | |
4P3T
 
 | |
4P43
 
 | |
2WPF
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00085762) | | Descriptor: | 3-[(4S)-6-CHLORO-2-METHYL-4-(4-METHYLPHENYL)QUINAZOLIN-3(4H)-YL]-N,N-DIMETHYLPROPAN-1-AMINE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
4P2C
 
 | | Complex of Shiga toxin 2e with a neutralizing single-domain antibody | | Descriptor: | Nanobody 1, Anti-F4+ETEC bacteria VHH variable region, Shiga toxin 2e, ... | | Authors: | Lo, A.W.H, Moonens, K, De Kerpel, M, Brys, L, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-03-03 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | The Molecular Mechanism of Shiga Toxin Stx2e Neutralization by a Single-domain Antibody Targeting the Cell Receptor-binding Domain.
J.Biol.Chem., 289, 2014
|
|
4P2E
 
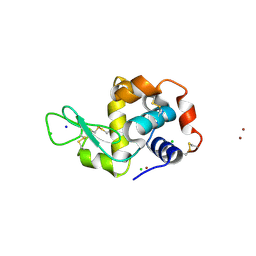 | | Acoustic transfer of protein crystals from agar pedestals to micromeshes for high throughput screening of heavy atom derivatives | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme C, ... | | Authors: | Cuttitta, C.M, Roessler, C.G, Ericson, D.L, Scalia, A, Teplitsky, E, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-03-03 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acoustic transfer of protein crystals from agarose pedestals to micromeshes for high-throughput screening.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4P7O
 
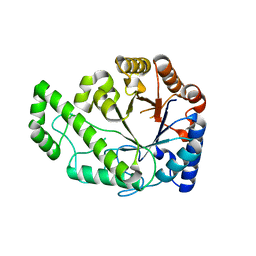 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8AGG
 
 | |
8ALK
 
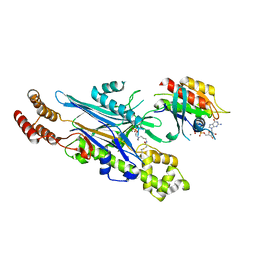 | | Structure of the Legionella phosphocholine hydrolase Lem3 in complex with its substrate Rab1 | | Descriptor: | 2-[[[5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxyethyl-[3-(2-chloranylethanoylamino)propyl]-dimethyl-azanium, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kaspers, M.S, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
4P2A
 
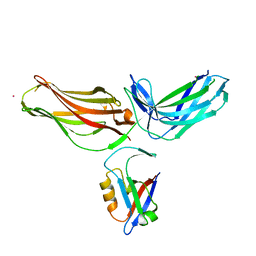 | | Structure of mouse VPS26A bound to rat SNX27 PDZ domain | | Descriptor: | MERCURY (II) ION, Sorting nexin-27, Vacuolar protein sorting-associated protein 26A | | Authors: | Clairfeuille, T, Gallon, M, Mas, C, Ghai, R, Teasdale, R, Cullen, P, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P9R
 
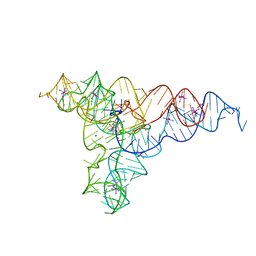 | | Speciation of a group I intron into a lariat capping ribozyme (Heavy atom derivative) | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (189-MER) | | Authors: | Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E, Masquida, B. | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Speciation of a group I intron into a lariat capping ribozyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3PVH
 
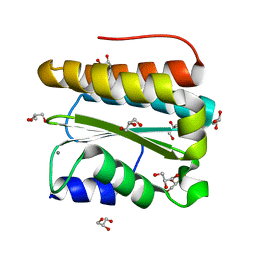 | | Structural and Functional Analysis of Arabidopsis thaliana thylakoid lumen protein AtTLP18.3 | | Descriptor: | CALCIUM ION, GLYCEROL, UPF0603 protein At1g54780, ... | | Authors: | Wu, H.Y, Liu, M.S, Lin, T.P, Cheng, Y.S. | | Deposit date: | 2010-12-07 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen
Plant Physiol., 157, 2011
|
|
4PA1
 
 | |
4P3O
 
 | | Structural Basis for Full-Spectrum Inhibition of Threonyl-tRNA Synthetase by Borrelidin 2 | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, GLYCEROL, Threonine--tRNA ligase, ... | | Authors: | Fang, P, Yu, X, Chen, K, Chen, X, Guo, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase.
Nat Commun, 6, 2015
|
|
4PAW
 
 | | Structure of hypothetical protein HP1257. | | Descriptor: | Orotate phosphoribosyltransferase, PHOSPHATE ION, THIOCYANATE ION, ... | | Authors: | Battaile, K.P, Lam, R, Lam, K, Romanov, V, Jones, K, Soloveychik, M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of hypothetical protein HP1257.
To Be Published
|
|
2WYO
 
 | | Trypanosoma brucei glutathione synthetase | | Descriptor: | GLUTATHIONE, GLUTATHIONE SYNTHETASE, SULFATE ION | | Authors: | Fyfe, P.K, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2009-11-17 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of Trypanosoma Brucei Glutathione Synthetase; Domain and Loop Alterations in the Catalytic Cycle of a Highly Conserved Enzyme.
Mol.Biochem.Parasitol., 170, 2010
|
|
