8HAH
 
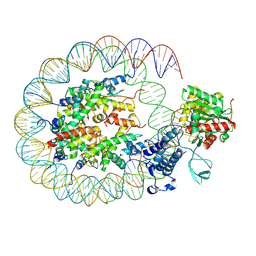 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAM
 
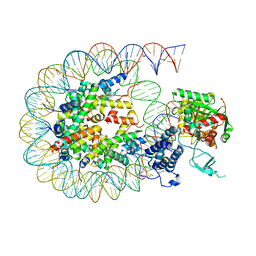 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 2 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAN
 
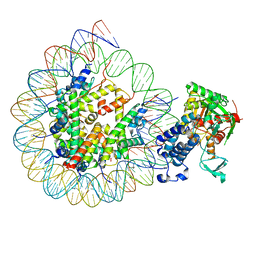 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
3QQQ
 
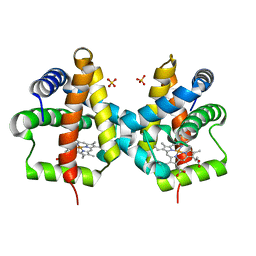 | | Crystal structure of non-symbiotic plant hemoglobin from Trema tomentosa | | 分子名称: | Non-symbiotic hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Kakar, S, Sturms, R, Savage, A, Nix, J.C, Dispirito, A, Hargrove, M.S. | | 登録日 | 2011-02-16 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal structures of Parasponia and Trema hemoglobins: differential heme coordination is linked to quaternary structure.
Biochemistry, 50, 2011
|
|
8HAK
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 4 (4.5 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
1LZN
 
 | | NEUTRON STRUCTURE OF HEN EGG-WHITE LYSOZYME | | 分子名称: | NITRATE ION, PROTEIN (LYSOZYME), SODIUM ION | | 著者 | Bon, C.I, Lehmann, M.S, Wilkinson, C. | | 登録日 | 1999-03-23 | | 公開日 | 1999-04-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å) | | 主引用文献 | Quasi-Laue neutron-diffraction study of the water arrangement in crystals of triclinic hen egg-white lysozyme.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4MPK
 
 | | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, GLYCEROL | | 著者 | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2013-09-13 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution
To be Published
|
|
4MPS
 
 | | Crystal structure of rat Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1), Northeast Structural Genomics Consortium Target RnR367A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactoside alpha-2,6-sialyltransferase 1 | | 著者 | Forouhar, F, Meng, L, Milaninia, S, Seetharaman, J, Su, M, Kornhaber, G, Montelione, G.T, Hunt, J.F, Moremen, K.W, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-09-13 | | 公開日 | 2013-09-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Enzymatic Basis for N-Glycan Sialylation: STRUCTURE OF RAT alpha 2,6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION.
J.Biol.Chem., 288, 2013
|
|
4MR9
 
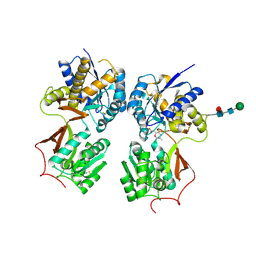 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist SCH50911 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | 著者 | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | 登録日 | 2013-09-17 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MS4
 
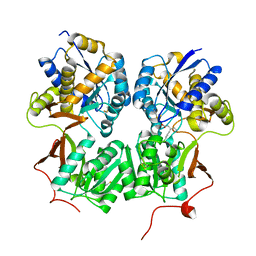 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the agonist baclofen | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | 著者 | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | 登録日 | 2013-09-18 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MQM
 
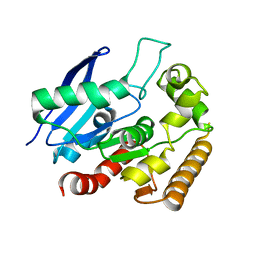 | | Crystal structure of Antigen 85C in presence of Ebselen | | 分子名称: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | 著者 | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | 登録日 | 2013-09-16 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.346 Å) | | 主引用文献 | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
3S2E
 
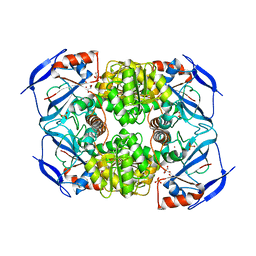 | | Crystal Structure of FurX NADH Complex 1 | | 分子名称: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | 登録日 | 2011-05-16 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.763 Å) | | 主引用文献 | Crystal Structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
4MVE
 
 | |
8FWI
 
 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2023-01-22 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWJ
 
 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2023-01-22 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
6NFA
 
 | |
3SIH
 
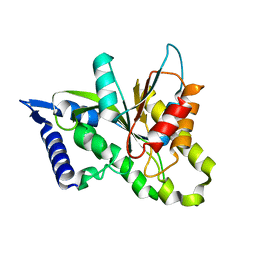 | |
4MZS
 
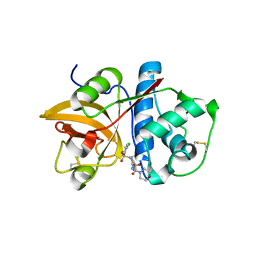 | |
4N01
 
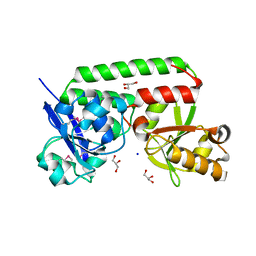 | |
4N1B
 
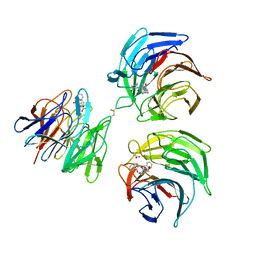 | | STRUCTURE OF KEAP1 KELCH DOMAIN WITH(1S,2R)-2-[(1S)-1-[(1-oxo-2,3-dihydro-1H-isoindol-2-Yl)methyl]-1,2,3,4-tetrahydroisoquinoline-2-Carbonyl]cyclohexane-1-carboxylic acid | | 分子名称: | (1S,2R)-2-{[(1S)-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | 著者 | Smith, M.A, Duclos, S, Beaumont, E, Kwong, J, Brooks, M, Barker, J, Jnoff, E, Brookfield, F, Courade, J.P, Barker, O, Fryatt, T, Albrecht, C, Bromidge, S. | | 登録日 | 2013-10-03 | | 公開日 | 2014-02-19 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4N2H
 
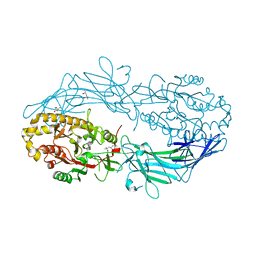 | | Crystal structure of Protein Arginine Deiminase 2 (D177A, 0 mM Ca2+) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | 著者 | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | 登録日 | 2013-10-04 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.808 Å) | | 主引用文献 | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N3E
 
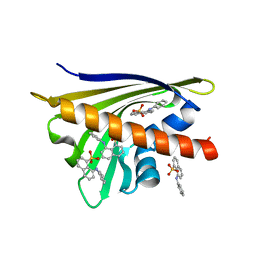 | | Crystal structure of Hyp-1, a St John's wort PR-10 protein, in complex with 8-anilino-1-naphthalene sulfonate (ANS) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, Phenolic oxidative coupling protein, ... | | 著者 | Sliwiak, J, Dauter, Z, Mccoy, A.J, Read, R.J, Jaskolski, M. | | 登録日 | 2013-10-07 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Likelihood-based molecular-replacement solution for a highly pathological crystal with tetartohedral twinning and sevenfold translational noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N0V
 
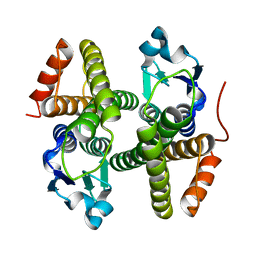 | | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332 | | 分子名称: | Glutathione S-transferase, N-terminal domain | | 著者 | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-10-02 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332
TO BE PUBLISHED
|
|
