1DK4
 
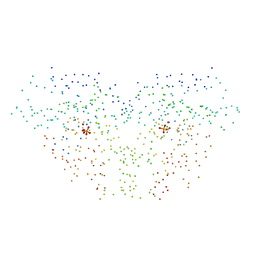 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
4HT3
 
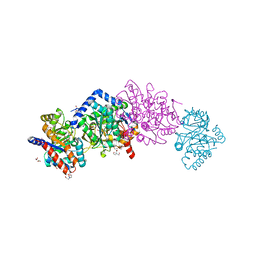 | | The crystal structure of Salmonella typhimurium Tryptophan Synthase at 1.30A complexed with N-(4'-TRIFLUOROMETHOXYBENZENESULFONYL)-2-AMINO-1-ETHYLPHOSPHATE (F9) inhibitor in the alpha site, internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-31 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
1F6U
 
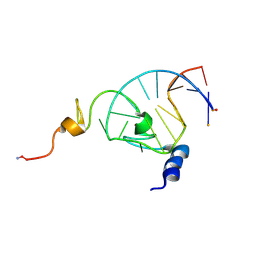 | | NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop sl2 of the psi-RNA packaging signal. Implications for genome recognition | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, HIV-1 STEM-LOOP SL2 FROM PSI-RNA PACKAGING, ZINC ION | | Authors: | Amarasinghe, G.K, De Guzman, R.N, Turner, R.B, Chancellor, K.J, Summers, M.F. | | Deposit date: | 2000-06-23 | | Release date: | 2000-10-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition.
J.Mol.Biol., 301, 2000
|
|
1EEA
 
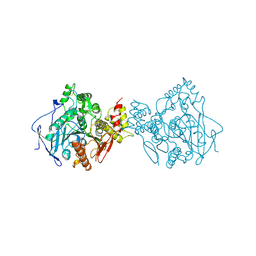 | | Acetylcholinesterase | | Descriptor: | PROTEIN (ACETYLCHOLINESTERASE) | | Authors: | Raves, M.L, Giles, K, Schrag, J.D, Schmid, M.F, Phillips Jr, G.N, Wah, C, Howard, A.J, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary Structure of Tetrameric Acetylcholinesterase
Structure and Function of Cholinesterases and Related Proteins, 1998
|
|
4HPX
 
 | | Crystal structure of Tryptophan Synthase at 1.65 A resolution in complex with alpha aminoacrylate E(A-A) and benzimidazole in the beta site and the F9 inhibitor in the alpha site | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
4I9J
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to diC4PC | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I8Y
 
 | | Structure of the unliganded N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I6E
 
 | | A vertebrate cryptochrome | | Descriptor: | Cryptochrome-2 | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4I6G
 
 | | a vertebrate cryptochrome with FAD | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4I90
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to choline | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9M
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus bound to HEPES | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4HPJ
 
 | | Crystal structure of Tryptophan Synthase at 1.45 A resolution in complex with 2-aminophenol quinonoid in the beta site and the F9 inhibitor in the alpha site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-23 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
4I9T
 
 | | Structure of the H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, SULFATE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
1Z1J
 
 | | Crystal structure of SARS 3CLpro C145A mutant | | Descriptor: | 3C-like proteinase | | Authors: | Hsu, M.F. | | Deposit date: | 2005-03-04 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Understanding the maturation process and inhibitor design of SARS-CoV 3CLpro from the crystal structure of C145A in a product-bound form
J.Biol.Chem., 280, 2005
|
|
2A3A
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with theophylline | | Descriptor: | SULFATE ION, THEOPHYLLINE, chitinase | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
2QFL
 
 | | Structure of SuhB: Inositol monophosphatase and extragenic suppressor from E. coli | | Descriptor: | ACETATE ION, ETHYL ACETATE, Inositol-1-monophosphatase | | Authors: | Wang, Y, Stieglitz, K.A, Bubunenko, M, Court, D, Stec, B, Roberts, M.F. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the R184A mutant of the inositol monophosphatase encoded by suhB and implications for its functional interactions in Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
2A3E
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, SULFATE ION, ... | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
2A75
 
 | | Trypanosoma rangeli Sialidase In Complex With 2,3- Difluorosialic Acid (Covalent Intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, SULFATE ION, sialidase | | Authors: | Amaya, M.F, Alzari, P.M, Buschiazzo, A. | | Deposit date: | 2005-07-04 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Analysis of Two Covalent Sialosyl-Enzyme Intermediates on Trypanosoma rangeli Sialidase.
J.Biol.Chem., 281, 2006
|
|
2QFK
 
 | | X-ray Crystal Structure Analysis of the Binding Site in the Ferric and Oxyferrous Forms of the Recombinant Heme Dehaloperoxidase Cloned from Amphitrite ornata | | Descriptor: | AMMONIUM ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, Chen, Z, Davis, M.F, Franzen, S. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structural analysis of the binding site in the ferric and
oxyferrous forms of the recombinant heme dehaloperoxidase cloned from Amphitrite ornata
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2A3B
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with caffeine | | Descriptor: | CAFFEINE, SULFATE ION, chitinase | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
2ATX
 
 | | Crystal Structure of the TC10 GppNHp complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, small GTP binding protein TC10 | | Authors: | Hemsath, L, Dvorsky, R, Fiegen, D, Carlier, M.F, Ahmadian, M.R. | | Deposit date: | 2005-08-26 | | Release date: | 2005-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins
Mol.Cell, 20, 2005
|
|
2AGS
 
 | | Trypanosoma rangeli Sialidase in Complex with 2-Keto-3-deoxy-D-glycero-D-galacto-2,3-difluoro-nononic acid (2,3-difluoro-KDN) | | Descriptor: | 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, SULFATE ION, sialidase | | Authors: | Amaya, M.F, Alzari, P.M, Buschiazzo, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Kinetic Analysis of Two Covalent Sialosyl-Enzyme Intermediates on Trypanosoma rangeli Sialidase.
J.Biol.Chem., 281, 2006
|
|
2QFN
 
 | | X-ray Crystal Structure Analysis of the Binding Site in the Ferric and Oxyferrous Forms of the Recombinant Heme Dehaloperoxidase Cloned from Amphitrite ornata | | Descriptor: | AMMONIUM ION, Dehaloperoxidase A, OXYGEN MOLECULE, ... | | Authors: | de Serrano, V, Chen, Z, Davis, M.F, Franzen, S. | | Deposit date: | 2007-06-27 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structural analysis of the binding site in the ferric and oxyferrous forms of the recombinant heme dehaloperoxidase cloned from Amphitrite ornata
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1ZRP
 
 | | SOLUTION-STATE STRUCTURE BY NMR OF ZINC-SUBSTITUTED RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Blake, P.R, Park, J.B, Zhou, Z.H, Hare, D.R, Adams, M.W.W, Summers, M.F. | | Deposit date: | 1992-07-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure by NMR of zinc-substituted rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
