1HVO
 
 | |
1HJW
 
 | | Crystal structure of hcgp-39 in complex with chitin octamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
1UNR
 
 | | Crystal structure of the PH domain of PKB alpha in complex with a sulfate molecule | | Descriptor: | RAC-ALPHA SERINE/THREONINE KINASE, SULFATE ION | | Authors: | Milburn, C.C, Deak, M, Kelly, S.M, Price, N.C, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2003-09-15 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change.
Biochem. J., 375, 2003
|
|
1UPQ
 
 | | Crystal structure of the pleckstrin homology (PH) domain of PEPP1 | | Descriptor: | PEPP1, SULFATE ION | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
1UPK
 
 | | Crystal structure of MO25 in complex with a C-terminal peptide of STRAD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MO25 PROTEIN, STE-20 RELATED ADAPTOR | | Authors: | Milburn, C.C, Boudeau, J, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-07 | | Release date: | 2004-01-22 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Mo25 Alpha in Complex with the C-Terminus of the Pseudo Kinase Ste-20 Related Adaptor (Strad)
Nat.Struct.Mol.Biol., 11, 2004
|
|
2KW3
 
 | | Heterotrimeric interaction between RFX5 and RFXAP | | Descriptor: | DNA-binding protein RFX5, Regulatory factor X-associated protein | | Authors: | Laird, K.M, Briggs, L.L, Boss, J.M, Summers, M.F, Garvie, C.W. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the heterotrimeric complex between RFX5 and RFXAP
reveals molecular details associated with MHCII gene expression
To be Published
|
|
1UPH
 
 | | HIV-1 Myristoylated Matrix | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Loeliger, E, Luncsford, P, Kinde, I, Beckett, D, Summers, M.F. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Entropic Switch Regulates Myristate Exposure in the HIV-1 Matrix Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2L3L
 
 | | The solution structure of the N-terminal domain of human Tubulin Binding Cofactor C reveals a platform for the interaction with ab-tubulin | | Descriptor: | Tubulin-specific chaperone C | | Authors: | Garcia-Mayoral, M.F, Castano, R, Lopez-Fanarraga, M.L, Zabala, J.C, Rico, M, Bruix, M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of human tubulin binding cofactor C reveals a platform for tubulin interaction
To be Published
|
|
1H9G
 
 | | FadR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI, in complex with myristoyl-CoA | | Descriptor: | COENZYME A, FATTY ACID METABOLISM REGULATOR PROTEIN, MYRISTIC ACID | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
1H9T
 
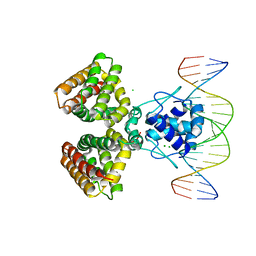 | | FADR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI IN COMPLEX WITH FADB OPERATOR | | Descriptor: | 5'-D(*CP*AP*TP*CP*TP*GP*GP*TP*AP*CP*GP*AP* CP*CP*AP*GP*AP*TP*C)-3', 5'-D(*GP*AP*TP*CP*TP*GP*GP*TP*CP*GP*TP*AP* CP*CP*AP*GP*AP*TP*G)-3', CHLORIDE ION, ... | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
1GSV
 
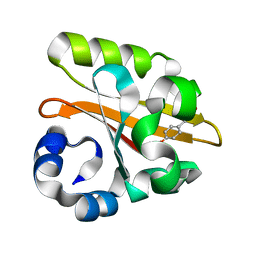 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1WWG
 
 | | NMR Structure Determined for MLV NC Complex with RNA Sequence UAUCUG | | Descriptor: | 5'-R(P*UP*AP*UP*CP*UP*G)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
1H10
 
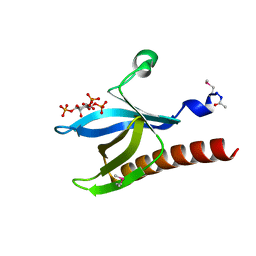 | | HIGH RESOLUTION STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN OF PROTEIN KINASE B/AKT BOUND TO INS(1,3,4,5)-TETRAKISPHOPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-ALPHA SERINE/THREONINE KINASE | | Authors: | Thomas, C.C, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Structure of the Pleckstrin Homology Domain of Protein Kinase B/Akt Bound to Phosphatidylinositol (3,4,5)-Trisphosphate
Curr.Biol., 12, 2002
|
|
1WWF
 
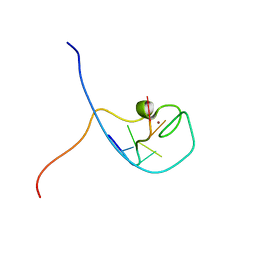 | | NMR Structure Determined for MLV NC Complex with RNA Sequence CCUCCGU | | Descriptor: | 5'-R(P*CP*CP*UP*CP*CP*GP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
1H0Y
 
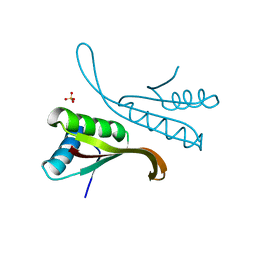 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B, SULFATE ION | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
1GWP
 
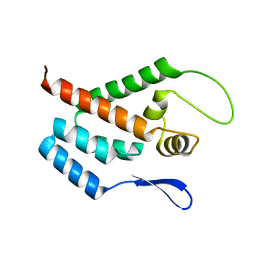 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE MATURE HIV-1 CAPSID PROTEIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Gitti, R.K, Lee, B.M, Walker, J, Summers, M.F, Yoo, S, Sundquist, W.I. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal 283-Residue Fragment of the Immature HIV-1 Gag Polyprotein
Nat.Struct.Biol., 9, 2002
|
|
2MDR
 
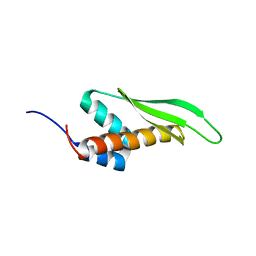 | | Solution structure of the third double-stranded RNA-binding domain (dsRBD3) of human adenosine-deaminase ADAR1 | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Barraud, P, Banerjee, S, Mohamed, W.I, Jantsch, M.F, Allain, F.H. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1H0X
 
 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
1WCH
 
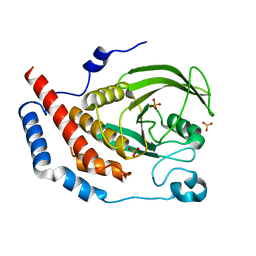 | | Crystal structure of PTPL1 human tyrosine phosphatase mutated in colorectal cancer - evidence for a second phosphotyrosine substrate recognition pocket | | Descriptor: | PHOSPHATE ION, PROTEIN TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 13 | | Authors: | Villa, F, Deak, M, Bloomberg, G.B, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Ptpl1/Fap-1 Human Tyrosine Phosphatase Mutated in Colorectal Cancer: Eveidence for a Second Phosphotyrosine Substrate Recognition Pocket
J.Biol.Chem., 280, 2005
|
|
1WCS
 
 | | A mutant of Trypanosoma rangeli sialidase displaying trans-sialidase activity | | Descriptor: | SIALIDASE | | Authors: | Paris, G, Ratier, L, Amaya, M.F, Nguyen, T, Alzari, P.M, Frasch, C. | | Deposit date: | 2004-11-21 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Sialidase Mutant Displaying Trans-Sialidase Activity
J.Mol.Biol., 345, 2005
|
|
2JKZ
 
 | | SACCHAROMYCES CEREVISIAE HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH GMP (GUANOSINE 5'- MONOPHOSPHATE) (ORTHORHOMBIC CRYSTAL FORM) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Moynie, L, Giraud, M.F, Breton, A, Boissier, F, Daignan-Fornier, B, Dautant, A. | | Deposit date: | 2008-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Protein Sci., 21, 2012
|
|
1H0I
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H0G
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argadin from Clonostachys | | Descriptor: | Argadin, CHITINASE B, GLYCEROL | | Authors: | Houston, D, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GSW
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSX
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G47S/G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
