1FOH
 
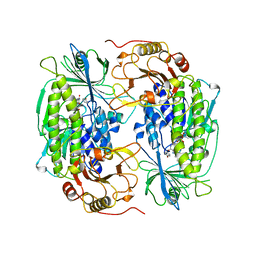 | | PHENOL HYDROXYLASE FROM TRICHOSPORON CUTANEUM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHENOL, PHENOL HYDROXYLASE | | Authors: | Enroth, C, Neujahr, H, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-03-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis.
Structure, 6, 1998
|
|
1B3N
 
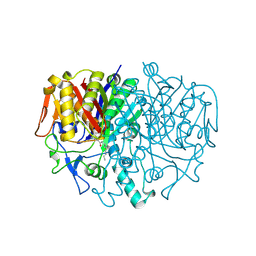 | | BETA-KETOACYL CARRIER PROTEIN SYNTHASE AS A DRUG TARGET, IMPLICATIONS FROM THE CRYSTAL STRUCTURE OF A COMPLEX WITH THE INHIBITOR CERULENIN. | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, PROTEIN (KETOACYL ACYL CARRIER PROTEIN SYNTHASE 2) | | Authors: | Moche, M, Schneider, G, Edwards, P, Dehesh, K, Lindqvist, Y. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase.
J.Biol.Chem., 274, 1999
|
|
1R0K
 
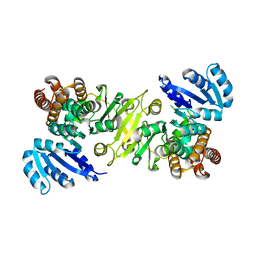 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Zymomonas mobilis | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ACETATE ION | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
1QZZ
 
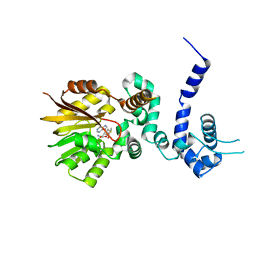 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1AY0
 
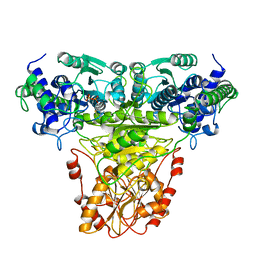 | | IDENTIFICATION OF CATALYTICALLY IMPORTANT RESIDUES IN YEAST TRANSKETOLASE | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, TRANSKETOLASE | | Authors: | Wikner, C, Nilsson, U, Meshalkina, L, Udekwu, C, Lindqvist, Y, Schneider, G. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytically important residues in yeast transketolase.
Biochemistry, 36, 1997
|
|
1R00
 
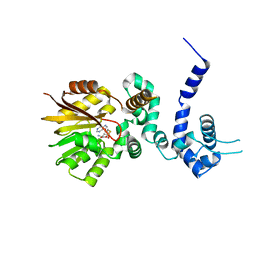 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-homocysteine (SAH) | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G. | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
2WG8
 
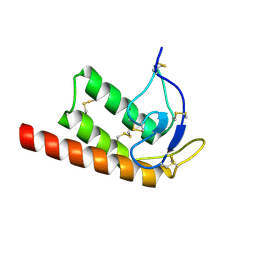 | | Structure of Oryza Sativa (Rice) PLA2, orthorhombic crystal form | | Descriptor: | CALCIUM ION, PUTATIVE PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Guy, J.E, Stahl, U, Lindqvist, Y. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Class Xib Phospholipase A2 (Pla2): Rice (Oryza Sativa) Isoform-2 Pla2 and an Octanoate Complex.
J.Biol.Chem., 284, 2009
|
|
1QJ3
 
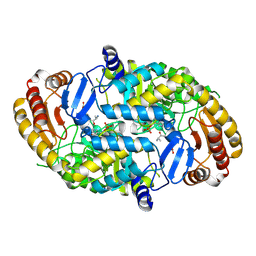 | | Crystal structure of 7,8-diaminopelargonic acid synthase in complex with 7-keto-8-aminopelargonic acid | | Descriptor: | 7,8-DIAMINOPELARGONIC ACID SYNTHASE, 7-KETO-8-AMINOPELARGONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Diaminopelargonic Acid Synthase; Evolutionary Relationships between Pyridoxal-5'-Phosphate Dependent Enzymes
J.Mol.Biol., 291, 1999
|
|
1R0L
 
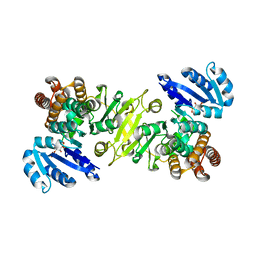 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from zymomonas mobilis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
1BR9
 
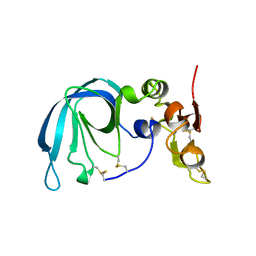 | | HUMAN TISSUE INHIBITOR OF METALLOPROTEINASE-2 | | Descriptor: | METALLOPROTEINASE-2 INHIBITOR | | Authors: | Tuuttila, A, Morgunova, E, Bergmann, U, Lindqvist, Y, Tryggvason, K, Schneider, G. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of human tissue inhibitor of metalloproteinases-2 at 2.1 A resolution.
J.Mol.Biol., 284, 1998
|
|
1H6V
 
 | | Mammalian thioredoxin reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | Authors: | Sandalova, T, Zhong, L, Lindqvist, Y, Holmgren, A, Schneider, G. | | Deposit date: | 2001-06-27 | | Release date: | 2001-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of a Mammalian Thioredoxin Reductase: Implication for Mechanism and Evolution of a Selenocysteine Dependent Enzyme
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1R7H
 
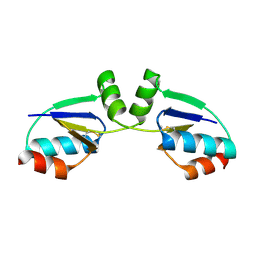 | |
1H7W
 
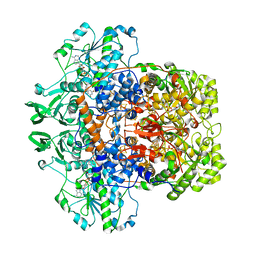 | | Dihydropyrimidine dehydrogenase (DPD) from pig | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
EMBO J., 20, 2001
|
|
1GT8
 
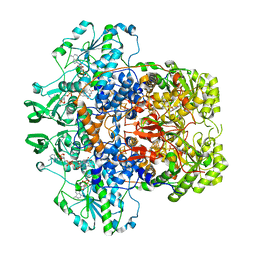 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
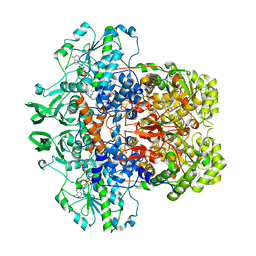 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTH
 
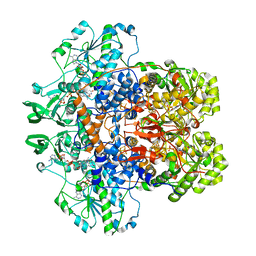 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1DAK
 
 | | DETHIOBIOTIN SYNTHETASE FROM ESCHERICHIA COLI, COMPLEX REACTION INTERMEDIATE ADP AND MIXED ANHYDRIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DETHIOBIOTIN SYNTHETASE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshot of a phosphorylated substrate intermediate by kinetic crystallography.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1H75
 
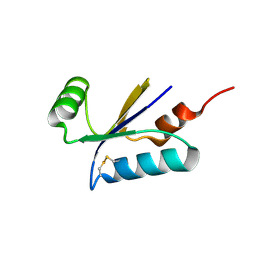 | | Structural basis for the thioredoxin-like activity profile of the glutaredoxin-like protein NrdH-redoxin from Escherichia coli. | | Descriptor: | GLUTAREDOXIN-LIKE PROTEIN NRDH | | Authors: | Stehr, M, Schneider, G, Aslund, F, Holmgren, A, Lindqvist, Y. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-09 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Thioredoxin-Like Activity Profile of the Glutaredoxin-Like Nrdh-Redoxin from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1GV9
 
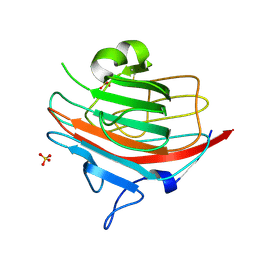 | | p58/ERGIC-53 | | Descriptor: | P58/ERGIC-53, SULFATE ION | | Authors: | Velloso, L.M, Svensson, K, Schneider, G, Pettersson, R.F, Lindqvist, Y. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-28 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of P58/Ergic-53, a Protein Involved in Glycoprotein Export from the Endoplasmic Reticulum.
J.Biol.Chem., 277, 2002
|
|
1DD8
 
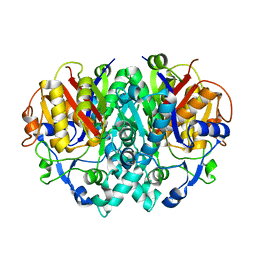 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I | | Authors: | Olsen, J.G, Kadziola, A, von Wettstein-Knowles, P, Siggaard-Andersen, M, Lindquist, Y, Larsen, S. | | Deposit date: | 1999-11-09 | | Release date: | 1999-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structure of beta-ketoacyl [acyl carrier protein] synthase I.
FEBS Lett., 460, 1999
|
|
1H7X
 
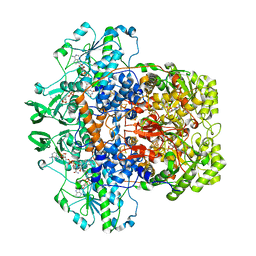 | | Dihydropyrimidine dehydrogenase (DPD) from pig, ternary complex of a mutant enzyme (C671A), NADPH and 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Dihydropyrimidine Dehydrogenase, a Major Determinant of the Pharmacokinetics of the Anti-Cancer Drug 5-Fluorouracil
Embo J., 20, 2001
|
|
1DTS
 
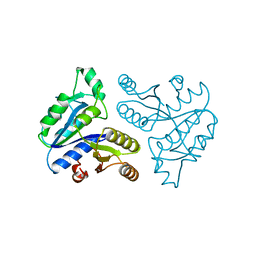 | | CRYSTAL STRUCTURE OF AN ATP DEPENDENT CARBOXYLASE, DETHIOBIOTIN SYNTHASE, AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-03-28 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an ATP-dependent carboxylase, dethiobiotin synthetase, at 1.65 A resolution.
Structure, 2, 1994
|
|
1CK7
 
 | | GELATINASE A (FULL-LENGTH) | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (GELATINASE A), ... | | Authors: | Morgunova, E, Tuuttila, A, Bergmann, U, Isupov, M, Lindqvist, Y, Schneider, G, Tryggvason, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed.
Science, 284, 1999
|
|
1R1Z
 
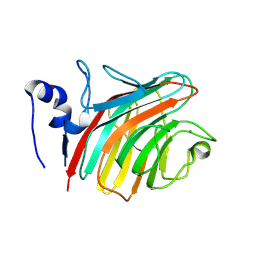 | | The Crystal structure of the Carbohydrate recognition domain of the glycoprotein sorting receptor p58/ERGIC-53 reveals a novel metal binding site and conformational changes associated with calcium ion binding | | Descriptor: | CALCIUM ION, ERGIC-53 protein | | Authors: | Velloso, L.M, Svensson, K, Pettersson, R.F, Lindqvist, Y. | | Deposit date: | 2003-09-25 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Carbohydrate-recognition Domain of the Glycoprotein Sorting Receptor p58/ERGIC-53 Reveals an Unpredicted Metal-binding Site and Conformational Changes Associated with Calcium Ion Binding.
J.Mol.Biol., 334, 2003
|
|
1R9J
 
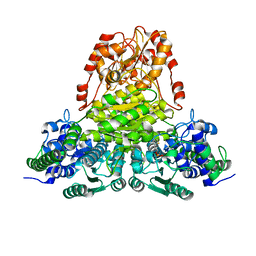 | | Transketolase from Leishmania mexicana | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, transketolase | | Authors: | Veitch, N.J, Mauger, D.A, Cazzulo, J.J, Lindqvist, Y, Barrett, M.P. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Transketolase from Leishmania mexicana has a dual subcellular localization.
Biochem.J., 382, 2004
|
|
