2GHW
 
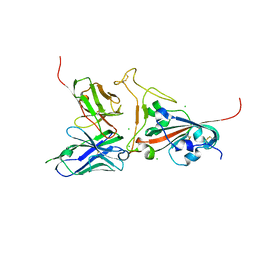 | | Crystal structure of SARS spike protein receptor binding domain in complex with a neutralizing antibody, 80R | | 分子名称: | CHLORIDE ION, Spike glycoprotein, anti-sars scFv antibody, ... | | 著者 | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | 登録日 | 2006-03-27 | | 公開日 | 2006-09-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2GHV
 
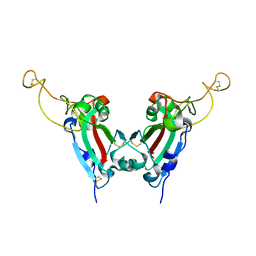 | | Crystal structure of SARS spike protein receptor binding domain | | 分子名称: | Spike glycoprotein | | 著者 | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | 登録日 | 2006-03-27 | | 公開日 | 2006-09-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
1KOF
 
 | | Crystal structure of gluconate kinase | | 分子名称: | Gluconate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | 登録日 | 2001-12-20 | | 公開日 | 2002-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KO4
 
 | |
1KO5
 
 | |
1KO8
 
 | |
1KNQ
 
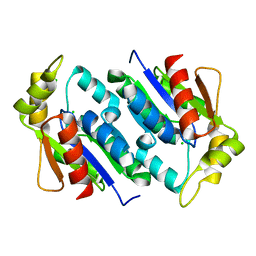 | |
1UCW
 
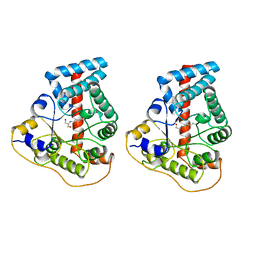 | |
4MDR
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain D190A mutant, in complex with a sorting peptide from the amyloid precursor protein (APP) | | 分子名称: | AP-4 complex subunit mu-1, Amyloid beta A4 protein | | 著者 | Ross, B.H, Lin, Y, Corales, E.A, Burgos, P.V, Mardones, G.A. | | 登録日 | 2013-08-23 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Functional Characterization of Cargo-Binding Sites on the mu 4-Subunit of Adaptor Protein Complex 4.
Plos One, 9, 2014
|
|
7Y62
 
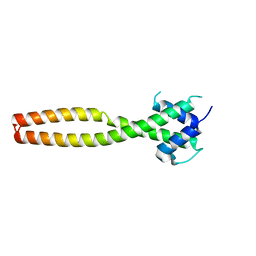 | | Crystal structure of human TFEB HLHLZ domain | | 分子名称: | Transcription factor EB | | 著者 | Yang, G, Li, P, Lin, Y, Liu, Z, Sun, H, Zhao, Z, Fang, P, Wang, J. | | 登録日 | 2022-06-18 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A small-molecule drug inhibits autophagy gene expression through the central regulator TFEB.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2LKN
 
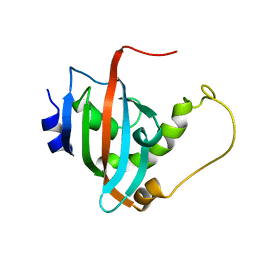 | | Solution structure of the PPIase domain of human aryl-hydrocarbon receptor-interacting protein (AIP) | | 分子名称: | AH receptor-interacting protein | | 著者 | Linnert, M, Lin, Y, Manns, A, Haupt, K, Paschke, A, Fischer, G, Weiwad, M, Luecke, C. | | 登録日 | 2011-10-17 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The FKBP-Type Domain of the Human Aryl Hydrocarbon Receptor-Interacting Protein Reveals an Unusual Hsp90 Interaction.
Biochemistry, 52, 2013
|
|
1BH0
 
 | | STRUCTURE OF A GLUCAGON ANALOG | | 分子名称: | GLUCAGON | | 著者 | Sturm, N.S, Lin, Y, Burley, S.K, Krstenansky, J.L, Ahn, J.-M, Azizeh, B.Y, Trivedi, D, Hruby, V.J. | | 登録日 | 1998-06-11 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-function studies on positions 17, 18, and 21 replacement analogues of glucagon: the importance of charged residues and salt bridges in glucagon biological activity.
J.Med.Chem., 41, 1998
|
|
1DUJ
 
 | | SOLUTION STRUCTURE OF THE SPINDLE ASSEMBLY CHECKPOINT PROTEIN HUMAN MAD2 | | 分子名称: | SPINDLE ASSEMBLY CHECKPOINT PROTEIN | | 著者 | Luo, X, Fang, G, Coldiron, M, Lin, Y, Yu, H. | | 登録日 | 2000-01-17 | | 公開日 | 2000-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20.
Nat.Struct.Biol., 7, 2000
|
|
1RHT
 
 | | 24-MER RNA HAIRPIN COAT PROTEIN BINDING SITE FOR BACTERIOPHAGE R17 (NMR, MINIMIZED AVERAGE STRUCTURE) | | 分子名称: | RNA (5'-R(P*GP*GP*GP*AP*CP*UP*GP*AP*CP*GP*AP*UP*CP*AP*CP*GP*CP*AP*GP*UP*CP*UP*AP*U)-3') | | 著者 | Borer, P.N, Lin, Y, Wang, S, Roggenbuck, M.W, Gott, J.M, Uhlenbeck, O.C, Pelczer, I. | | 登録日 | 1995-03-03 | | 公開日 | 1995-06-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Proton NMR and structural features of a 24-nucleotide RNA hairpin.
Biochemistry, 34, 1995
|
|
4V0J
 
 | | The channel-block Ser202Glu, Thr104Lys double mutant of Stearoyl-ACP- Desaturase from Castor bean (Ricinus communis) | | 分子名称: | ACYL-[ACYL-CARRIER-PROTEIN] DESATURASE, CHLOROPLASTIC, FE (II) ION, ... | | 著者 | Moche, M, Guy, J, Whittle, E, Lindqvist, Y, Shanklin, J. | | 登録日 | 2014-09-17 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Half-of-the-Sites Reactivity of the Castor Delta9-18:0-Acp Desaturase.
Plant Physiol., 169, 2015
|
|
2MOG
 
 | | Solution structure of the terminal Ig-like domain from Leptospira interrogans LigB | | 分子名称: | Bacterial Ig-like domain, group 2 | | 著者 | Ptak, C.P, Hsieh, C, Lin, Y, Maltsev, A.S, Raman, R, Sharma, Y, Oswald, R.E, Chang, Y. | | 登録日 | 2014-04-25 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Solution Structure of the Terminal Immunoglobulin-like Domain from the Leptospira Host-Interacting Outer Membrane Protein, LigB.
Biochemistry, 53, 2014
|
|
1I7K
 
 | |
2OEY
 
 | | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA | | 分子名称: | (1R,3A'S,10'S,10A'R)-7-METHOXY-2-OXO-10',10A'-DIHYDRO-2H,3A'H-SPIRO[NAPHTHALENE-1,3'-PENTALENO[1,2-B]NAPHTHALEN]-10'-YL 2,6-DIDEOXY-2-(METHYLAMINO)-ALPHA-D-GALACTOPYRANOSIDE, DNA (25-MER) | | 著者 | Zhang, N, Lin, Y, Xiao, Z, Jones, G.B, Goldberg, I.H. | | 登録日 | 2007-01-01 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA.
Biochemistry, 46, 2007
|
|
2N76
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | 分子名称: | De novo designed protein LFR1 | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-09-03 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414
To be Published
|
|
2N75
 
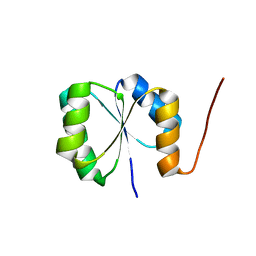 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | 分子名称: | De novo designed protein | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-09-03 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2N3Z
 
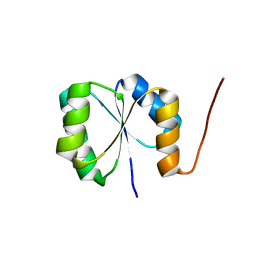 | | Solution NMR Structure of de novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | 分子名称: | OR446 | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-06-15 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2MQ8
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | 分子名称: | De novo designed protein LFR1 | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-06-12 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Control over overall shape and size in de novo designed proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N2T
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303 | | 分子名称: | OR303 | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-05-14 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303
To be Published
|
|
2N2U
 
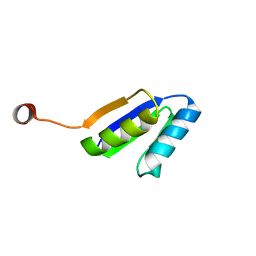 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | 分子名称: | OR358 | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-05-14 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
7XM8
 
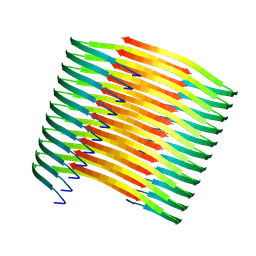 | |
