2V2U
 
 | | Structure of Mouse gammaC-crystallin | | Descriptor: | GAMMA CRYSTALLIN C | | Authors: | Purkiss, A.G, Bateman, O.A, Wyatt, K, David, L.L, Wistow, G.J, Slingsby, C. | | Deposit date: | 2007-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolutionary Adaptation of Human and Mouse Eye Lens Gammac-Crystallins for Solubility and Stability: Fine-Tuning of Molecular Dipoles.
J.Mol.Biol., 372, 2007
|
|
2BV2
 
 | | beta gamma crystallin from Ciona Intestinalis | | Descriptor: | ACETATE ION, CALCIUM ION, CIONA BETAGAMMA-CRYSTALLIN, ... | | Authors: | Shimeld, S.M, Purkiss, A.G, Dirks, R.P.H, Bateman, O.A, Slingsby, C, Lubsen, N.H. | | Deposit date: | 2005-06-21 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Urochordate Betagamma-Crystallin and the Evolutionary Origin of the Vertebrate Eye Lens.
Curr.Biol., 15, 2005
|
|
3DVQ
 
 | |
3DW1
 
 | |
2BOL
 
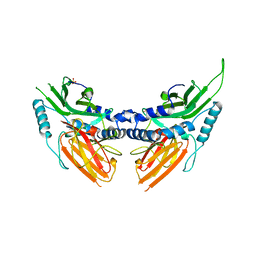 | | CRYSTAL STRUCTURE AND ASSEMBLY OF TSP36, A METAZOAN SMALL HEAT SHOCK PROTEIN | | Descriptor: | SMALL HEAT SHOCK PROTEIN, SULFATE ION | | Authors: | Stamler, R.J, Kappe, G, Boelens, W.C, Slingsby, C. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Wrapping the Alpha-Crystallin Domain Fold in a Chaperone Assembly.
J.Mol.Biol., 353, 2005
|
|
3V0I
 
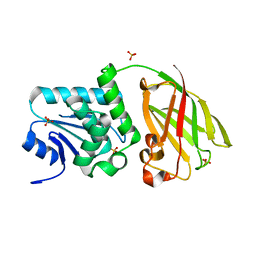 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 256-576, E411F | | Descriptor: | SULFATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2HSG
 
 | |
3V0J
 
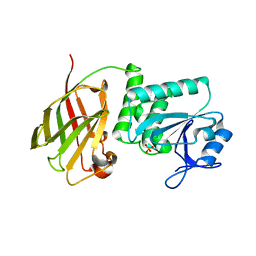 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), Deletion of 401-405 | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0G
 
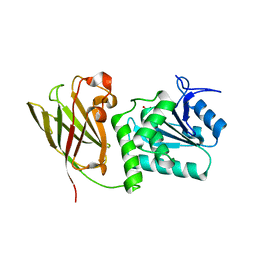 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), form III | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3DZN
 
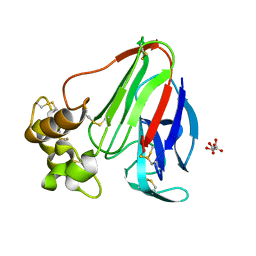 | |
3E0A
 
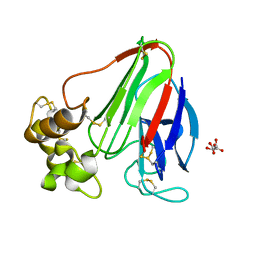 | |
2A54
 
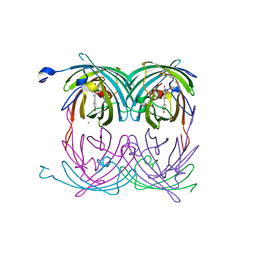 | | fluorescent protein asFP595, A143S, on-state, 1min irradiation | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3DE3
 
 | |
2AUB
 
 | | Lysozyme structure derived from thin-film-based crystals | | Descriptor: | Lysozyme C | | Authors: | Pechkova, E, Sivozhelezov, V, Tropiano, G, Fiordoro, S, Nicolini, C. | | Deposit date: | 2005-08-27 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of lysozyme structures derived from thin-film-based and classical crystals.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3DE5
 
 | |
3DO1
 
 | |
3DO0
 
 | |
3DVR
 
 | |
3DWE
 
 | |
4HR1
 
 | | Structure of PAV1-137, a protein from the virus PAV1 that infects Pyrococcus abyssi. | | Descriptor: | Putative uncharacterized protein | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, Geslin, C, Flament, D, Romancer, M.L, Tilbeurgh, H.V. | | Deposit date: | 2012-10-26 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of PAV1-137: a protein from the virus PAV1 that infects Pyrococcus abyssi
Archaea, 2013, 2013
|
|
3DNZ
 
 | |
3DE7
 
 | |
3DE4
 
 | |
3DVS
 
 | |
2A56
 
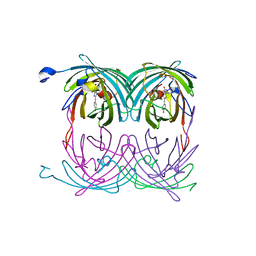 | | fluorescent protein asFP595, A143S, on-state, 5min irradiation | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
