1MNG
 
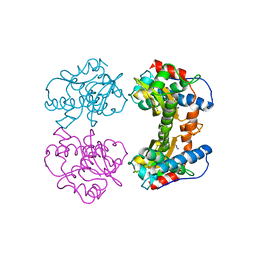 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | AZIDE ION, MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
1ISB
 
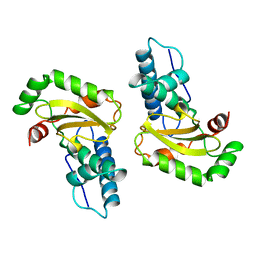 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | FE (III) ION, IRON(III) SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
1ISC
 
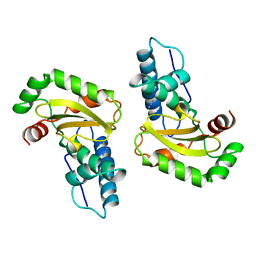 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | AZIDE ION, FE (III) ION, IRON(III) SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
1ISA
 
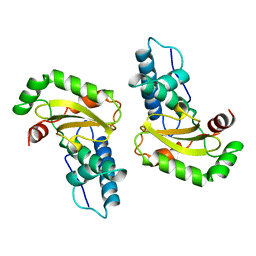 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | FE (II) ION, IRON(II) SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
1PXA
 
 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
1PXC
 
 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
1PXB
 
 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
8C8A
 
 | | The NMR structure of the MAX28 effector from Magnaporthe oryzae | | Descriptor: | But2 domain-containing protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Kroj, T, Roumestand, C, Barthe, P. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-14 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N Backbone and side-chain NMR assignments for a MAX effector from Magnaporthe oryzae
To Be Published
|
|
2VEK
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | 3-(BUTYLSULPHONYL)-PROPANOIC ACID, CITRIC ACID, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEN
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | CITRIC ACID, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEI
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEL
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties. | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEM
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | (3-bromo-2-oxo-propoxy)phosphonic acid, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2V0T
 
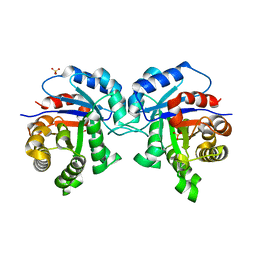 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-05-18 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies show that the A178L mutation in the C-terminal hinge of the catalytic loop-6 of triosephosphate isomerase (TIM) induces a closed-like conformation in dimeric and monomeric TIM.
Acta Crystallogr. D Biol. Crystallogr., 64, 2008
|
|
2V2H
 
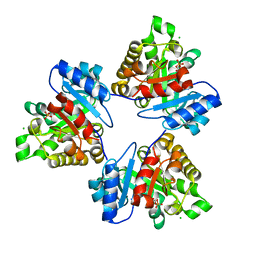 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-06-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed-Like Conformation in Dimeric and Monomeric Tim.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2V2C
 
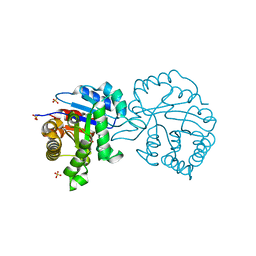 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed-Like Conformation in Dimeric and Monomeric Tim.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2V2D
 
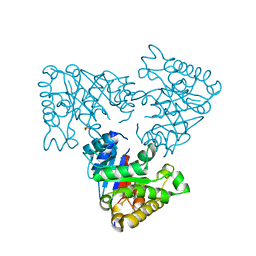 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | PHOSPHATE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed- Like Conformation in Dimeric and Monomeric Tim.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2VXN
 
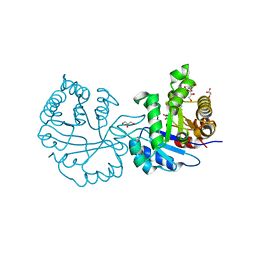 | |
7ZJY
 
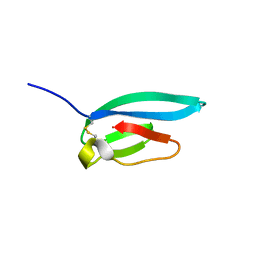 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
7ZK0
 
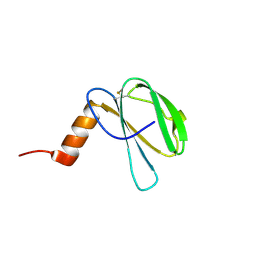 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
7ZKD
 
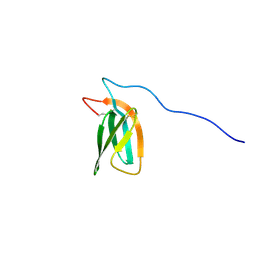 | | The NMR structure of the MAX47 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
3SK0
 
 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2011-06-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
4FWB
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 in complex with 1, 2, 3 - trichloropropane | | Descriptor: | 1,2,3-trichloropropane, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Kuta Smatanova, I. | | Deposit date: | 2012-06-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3RK4
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WE1
 
 | | Crystal structure of Dengue 4 Envelope protein domain III (ED3) | | Descriptor: | Envelope protein E | | Authors: | Elahi, M, Islam, M.M, Noguchi, K, Yohda, M, Toh, H, Kuroda, Y. | | Deposit date: | 2013-06-27 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Computational prediction and experimental characterization of a "size switch type repacking" during the evolution of dengue envelope protein domain III (ED3).
Biochim.Biophys.Acta, 1844, 2014
|
|
