6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | 分子名称: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | 著者 | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | 登録日 | 2017-09-15 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3UA3
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 in complex with SAH | | 分子名称: | Protein arginine N-methyltransferase 5, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | 登録日 | 2011-10-20 | | 公開日 | 2011-12-14 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | 分子名称: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | 著者 | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | 登録日 | 2013-11-27 | | 公開日 | 2014-11-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QP0
 
 | | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei | | 分子名称: | Endo-beta-mannanase, SULFATE ION | | 著者 | Zheng, Q.J, Peng, Z, Liu, Y, Yan, Q.J, Chen, Z.Z, Qin, Z. | | 登録日 | 2014-06-22 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7E29
 
 | | Crystal Structure of Saccharomyces cerevisiae Ioc4 PWWP domain fused with MBP | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,ISWI one complex protein 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Li, J, Smolle, M, Liang, H, Liu, Y. | | 登録日 | 2021-02-05 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | H3K36 methylation and DNA-binding both promote Ioc4 recruitment and Isw1b remodeler function.
Nucleic Acids Res., 50, 2022
|
|
1WP8
 
 | | crystal structure of Hendra Virus fusion core | | 分子名称: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | 著者 | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | 登録日 | 2004-08-31 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of Nipah and Hendra virus fusion core proteins
FEBS J., 273, 2006
|
|
8TXQ
 
 | | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2 | | 分子名称: | CBH-4B Heavy chain, CBH-4B Light chain, envelope glycoprotein E2 | | 著者 | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | 登録日 | 2023-08-24 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
7D3X
 
 | |
7D3W
 
 | |
2LQ9
 
 | |
2JVC
 
 | |
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | 分子名称: | Heat shock protein beta-1 | | 著者 | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | 登録日 | 2015-06-03 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
2OIG
 
 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | 分子名称: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | 著者 | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | 登録日 | 2007-01-11 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2OIE
 
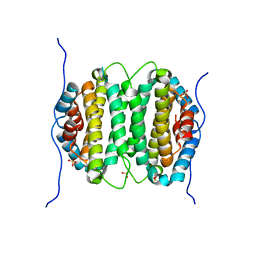 | | Crystal structure of RS21-C6 core segment RSCUT | | 分子名称: | RS21-C6, SULFATE ION | | 著者 | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | 登録日 | 2007-01-10 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2O0F
 
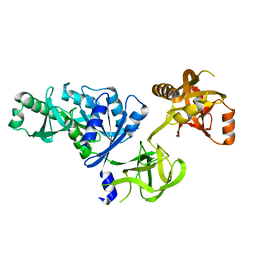 | | Docking of the modified RF3 X-ray structure into cryo-EM map of E.coli 70S ribosome bound with RF3 | | 分子名称: | Peptide chain release factor 3 | | 著者 | Gao, H, Zhou, Z, Rawat, U, Huang, C, Bouakaz, L, Wang, C, Liu, Y, Zavialov, A, Gursky, R, Sanyal, S, Ehrenberg, M, Frank, J, Song, H. | | 登録日 | 2006-11-27 | | 公開日 | 2007-07-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (15.5 Å) | | 主引用文献 | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
2K5U
 
 | |
7W7P
 
 | |
5YDE
 
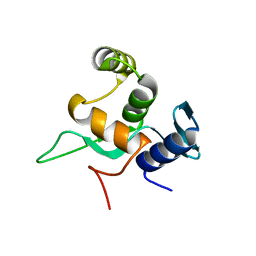 | | Crystal structure of a disease-related gene, hCDC73(1-111) | | 分子名称: | Parafibromin | | 著者 | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | 登録日 | 2017-09-13 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.023 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
5YDF
 
 | | Crystal structure of a disease-related gene, hCDC73(1-100) | | 分子名称: | Parafibromin | | 著者 | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | 登録日 | 2017-09-13 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
7XC4
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | 分子名称: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | 著者 | Li, J, Liu, Y, Gao, J, Ruan, K. | | 登録日 | 2022-03-22 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
5YY8
 
 | |
2K6S
 
 | |
8JIF
 
 | |
7WI7
 
 | |
7X8M
 
 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | 分子名称: | BERBERINE, DNA (24-MER) | | 著者 | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | 登録日 | 2022-03-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
