4LE4
 
 | | Crystal structure of PaGluc131A with cellotriose | | Descriptor: | Beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, T, Chan, H.C, Huang, C.H, Ko, T.P, Huang, T.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a GH131 beta-Glucanase Catalytic Domain from Podospora anserina in Complex with Cellotriose
To be Published
|
|
4ONC
 
 | | Crystal Structure of Mycobacterium Tuberculosis Decaprenyl Diphosphate Synthase in Complex with BPH-640 | | Descriptor: | Decaprenyl diphosphate synthase, [hydroxy(1,1':3',1''-terphenyl-3-yl)methanediyl]bis(phosphonic acid) | | Authors: | Feng, X, Chan, H.C, Ko, T.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Inhibition of Tuberculosinol Synthase and Decaprenyl Diphosphate Synthase from Mycobacterium tuberculosis
J.Am.Chem.Soc., 136, 2014
|
|
5H9D
 
 | | Crystal structure of Heptaprenyl Diphosphate Synthase from Staphylococcus aureus | | Descriptor: | C-terminal peptide from Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, Farnesyl pyrophosphate synthetase, Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, ... | | Authors: | Wei, H.L, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, Function, and Inhibition of Staphylococcus aureus Heptaprenyl Diphosphate Synthase
ChemMedChem, 11, 2016
|
|
4QNN
 
 | |
5XFT
 
 | |
6JD1
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADH, and CPD at pH7.5 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JD2
 
 | | Crystal structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+ at pH8.5 | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J.Am.Chem.Soc., 141, 2019
|
|
6JCW
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH8.5 | | Descriptor: | MAGNESIUM ION, ketol-acid reductoisomerase | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JCZ
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADPH, and CPD at pH7.5 | | Descriptor: | MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JU0
 
 | | Mouse antibody 3.3 Fab in complex with PEG | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Fab heavy chain, ... | | Authors: | Lee, C.C, Ko, T.P, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of polyethylene glycol recognition by antibody.
J.Biomed.Sci., 27, 2020
|
|
6JX7
 
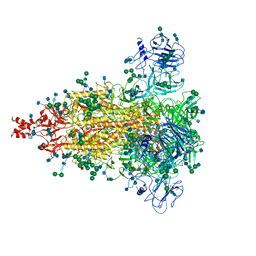 | | Cryo-EM structure of spike protein of feline infectious peritonitis virus strain UU4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Feline Infectious Peritonitis Virus Spike Protein, ... | | Authors: | Hsu, S.T.D, Yang, T.J, Ko, T.P, Draczkowski, P. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
6LYJ
 
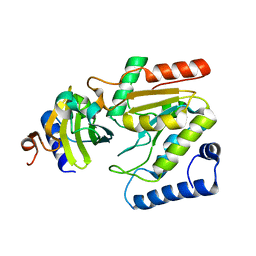 | | The crystal structure of SAUGI/EBVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
6LYV
 
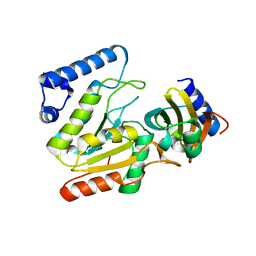 | | The crystal structure of SAUGI/KSHVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
6JWC
 
 | | Mouse antibody 2B5 Fab in complex with PEG | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, ... | | Authors: | Lee, C.C, Ko, T.P, Su, Y.C, Lin, L.L, Roffler, S.R, Wang, A.H.J. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural basis of polyethylene glycol recognition by antibody.
J.Biomed.Sci., 27, 2020
|
|
6M5N
 
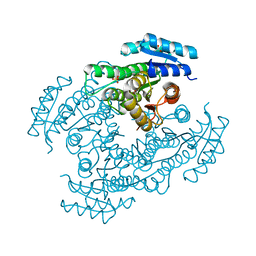 | | Apo-Form Structure of Borneol Dehydrogenase | | Descriptor: | ACETATE ION, Borneol dehydrogenase, SULFATE ION | | Authors: | Khine, A.A, Huang, K.F, Ko, T.P, Chen, H.P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural characterization of borneol dehydrogenase from Pseudomonas sp. TCU-HL1
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6JP7
 
 | | Human antibody 32D6 Fab in complex with PEG | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, HEXAETHYLENE GLYCOL, immunoglobulin Fab heavy chain, ... | | Authors: | Lee, C.C, Ko, T.P, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2019-03-26 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Structural basis of polyethylene glycol recognition by antibody.
J.Biomed.Sci., 27, 2020
|
|
6JCV
 
 | | Cryo-EM structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH7.5 | | Descriptor: | MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
2NZX
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase with GDP | | Descriptor: | Alpha1,3-fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
2NZW
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase | | Descriptor: | Alpha1,3-fucosyltransferase, SULFATE ION | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
2NZY
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose | | Descriptor: | Alpha1,3-Fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
1V4J
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima V73Y mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1V4H
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima F52A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1V4K
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima S77F mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1V4I
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima F132A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
