8GA6
 
 | |
8G9K
 
 | |
8FIH
 
 | |
8FIT
 
 | |
8FIN
 
 | |
8FJF
 
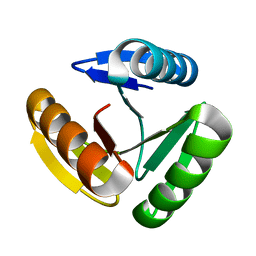 | | The three-repeat design H10 | | Descriptor: | H10 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FJE
 
 | | The five-repeat design E8 | | Descriptor: | E8 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FJG
 
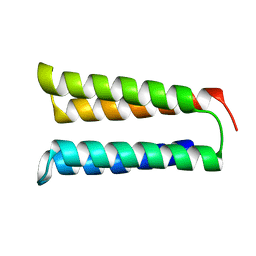 | | The two-repeat design H12 | | Descriptor: | H12 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DT0
 
 | |
8GJI
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | GCG binder, Glucagon | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GJG
 
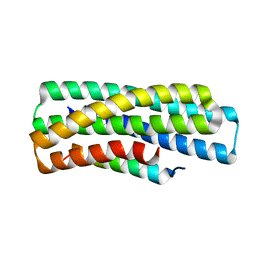 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | gluc_A04_0005, gluc_A04_0005 Binder | | Authors: | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GEL
 
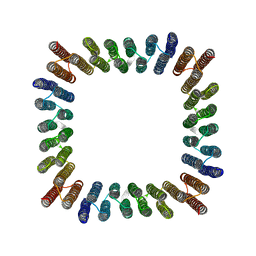 | | Cryo-EM structure of synthetic tetrameric building block sC4 | | Descriptor: | sC4 | | Authors: | Redler, R.L, Huddy, T.F, Hsia, Y, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Blueprinting extendable nanomaterials with standardized protein blocks.
Nature, 627, 2024
|
|
8FIQ
 
 | |
8FVT
 
 | |
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
8E55
 
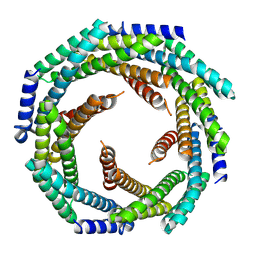 | |
8GLT
 
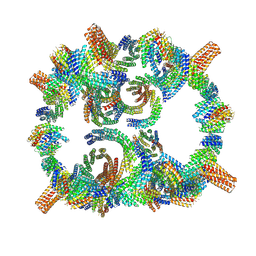 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | Descriptor: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | Authors: | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 20, 2024
|
|
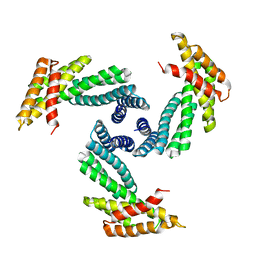
8E1E
 
 | |
6O35
 
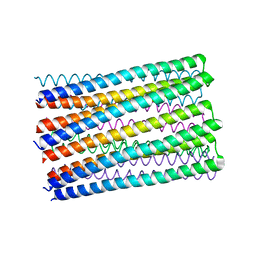 | |
8CYK
 
 | | Crystal structure of hallucinated protein HALC1_878 | | Descriptor: | HALC1_878 | | Authors: | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
8F54
 
 | |
8F4X
 
 | |
8F53
 
 | |
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8EJA
 
 | |
