6X90
 
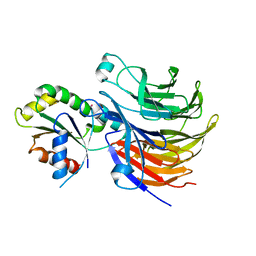 | |
7KMT
 
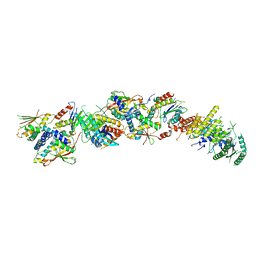 | | Structure of the yeast TRAPPIII-Ypt1(Rab1) complex | | 分子名称: | GTP-binding protein YPT1, PALMITIC ACID, Trafficking protein particle complex III-specific subunit 85, ... | | 著者 | Joiner, A.M.N, Phillips, B.P, Miller, E.A, Fromme, J.C. | | 登録日 | 2020-11-03 | | 公開日 | 2021-06-02 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of TRAPPIII-mediated Rab1 activation.
Embo J., 40, 2021
|
|
8GF5
 
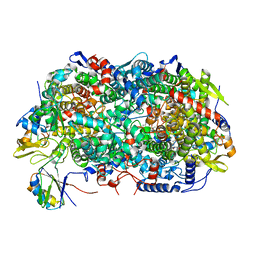 | | McrD binds asymmetrically to methyl-coenzyme M reductase improving active site accessibility during assembly | | 分子名称: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | 著者 | Joiner, A.M.N, Chadwick, G.L, Nayak, D.D. | | 登録日 | 2023-03-07 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | McrD binds asymmetrically to methyl-coenzyme M reductase improving active-site accessibility during assembly.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GF6
 
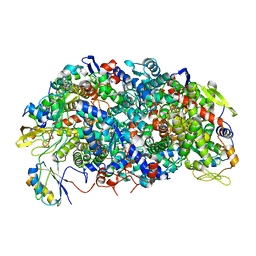 | | Apo-apo MCR assembly intermediate | | 分子名称: | Methyl coenzyme M reductase, subunit D, Methyl-coenzyme M reductase subunit alpha, ... | | 著者 | Joiner, A.M.N, Chadwick, G.L, Nayak, D.D. | | 登録日 | 2023-03-07 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | McrD binds asymmetrically to methyl-coenzyme M reductase improving active-site accessibility during assembly.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
