6ZVX
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
2W6X
 
 | | Crystal structure of Sperm Whale Myoglobin mutant YQRF in complex with Xenon | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Miele, A.E, Draghi, F, Renzi, F, Sciara, G, Johnson, K.A, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | When the Same Fold Does not Mean the Same Function: The Case of Xenon Cavities in Hemoglobin and Myoglobin
To be Published
|
|
2W6Y
 
 | | Crystal structure of Sperm Whale Myoglobin mutant YQR in complex with Xenon | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Miele, A.E, Draghi, F, Renzi, F, Sciara, G, Johnson, K.A, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | When the Same Fold Does not Mean the Same Function: The Case of Xenon Cavities in Hemoglobin and Myoglobin
To be Published
|
|
2X5F
 
 | | Crystal structure of the methicillin-resistant Staphylococcus aureus Sar2028, an aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate dependent aminotransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASPARTATE_TYROSINE_PHENYLALANINE PYRIDOXAL-5' PHOSPHATE-DEPENDENT AMINOTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
1GJY
 
 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
3OHT
 
 | | Crystal Structure of Salmo Salar p38alpha | | Descriptor: | N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, SULFATE ION, p38a | | Authors: | Rothweiler, U, Johnson, K, Engh, R.A. | | Deposit date: | 2010-08-18 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | p38alpha MAP kinase dimers with swapped activation segments and a novel catalytic loop conformation
J.Mol.Biol., 411, 2011
|
|
2X5D
 
 | | Crystal Structure of a probable aminotransferase from Pseudomonas aeruginosa | | Descriptor: | PROBABLE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2VH3
 
 | | ranasmurfin | | Descriptor: | GLYCEROL, RANASMURFIN, SULFATE ION, ... | | Authors: | Oke, M, Ching, R.T, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, Bloch Junior, C, Botting, C.H, Walsh, M.A, Latiff, A.A, Kennedy, M.W, Cooper, A, Naismith, J.H. | | Deposit date: | 2007-11-17 | | Release date: | 2007-12-04 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Unusual Chromophore and Cross-Links in Ranasmurfin: A Blue Protein from the Foam Nests of a Tropical Frog.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
2VHS
 
 | | Cathsilicatein, a chimera | | Descriptor: | CATHSILICATEIN, SULFATE ION | | Authors: | Fairhead, M, Kowatz, T, McMahon, S.A, Carter, L.G, Oke, M, Johnson, K.A, Liu, H, Naismith, J.H, Wal, C.F.V.D. | | Deposit date: | 2007-11-24 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Silica Condensing Activities of Silicatein Alpha-Cathepsin L Chimeras.
Chem. Commun., 2008
|
|
3L00
 
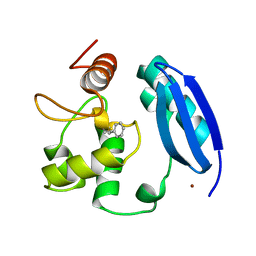 | |
3KZY
 
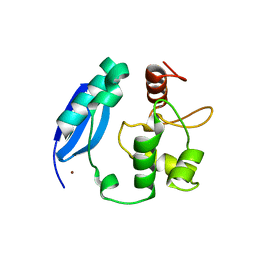 | | Crystal structure of SNAP-tag | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, ZINC ION | | Authors: | Bannwarth, M, Schmitt, S, Pojer, F, Schiltz, M, Johnsson, K. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | SNAP-tag structure
To be Published
|
|
3KZZ
 
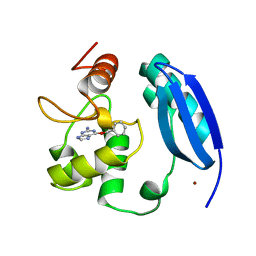 | |
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
3FFE
 
 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2W6V
 
 | | Structure of Human deoxy Hemoglobin A in complex with Xenon | | Descriptor: | GLYCEROL, HEMOGLOBIN SUBUNIT ALPHA, HEMOGLOBIN SUBUNIT BETA, ... | | Authors: | Miele, A.E, Draghi, F, Sciara, G, Johnson, K.A, Renzi, F, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pattern of Cavities in Globins: The Case of Human Hemoglobin.
Biopolymers, 91, 2009
|
|
2X3L
 
 | | Crystal Structure of the Orn_Lys_Arg decarboxylase family protein SAR0482 from Methicillin-resistant Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, ORN/LYS/ARG DECARBOXYLASE FAMILY PROTEIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1S5J
 
 | | Insight in DNA Replication: The crystal structure of DNA Polymerase B1 from the archaeon Sulfolobus solfataricus | | Descriptor: | DNA polymerase I, MAGNESIUM ION, SULFATE ION | | Authors: | Savino, C, Federici, L, Nastopoulos, V, Johnson, K.A, Pisani, F.M, Rossi, M, Tsernoglou, D. | | Deposit date: | 2004-01-21 | | Release date: | 2004-11-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into DNA replication: the crystal structure of DNA polymerase B1 from the archaeon Sulfolobus solfataricus
Structure, 12, 2004
|
|
2X4I
 
 | | ORF 114a from Sulfolobus islandicus rudivirus 1 | | Descriptor: | UNCHARACTERIZED PROTEIN 114 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-31 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4K
 
 | | Crystal structure of SAR1376, a putative 4-oxalocrotonate tautomerase from the methicillin-resistant Staphylococcus aureus (MRSA) | | Descriptor: | 4-OXALOCROTONATE TAUTOMERASE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4H
 
 | | Crystal Structure of the hypothetical protein SSo2273 from Sulfolobus solfataricus | | Descriptor: | HYPOTHETICAL PROTEIN SSO2273, ZINC ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-31 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3D
 
 | | Crystal Structure of SSo6206 from Sulfolobus solfataricus P2 | | Descriptor: | SSO6206 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, McEwan, A.R, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: targets, methods and outputs.
J. Struct. Funct. Genomics, 11, 2010
|
|
2X3F
 
 | | Crystal Structure of the Methicillin-Resistant Staphylococcus aureus Sar2676, a Pantothenate Synthetase. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PANTHOTHENATE SYNTHETASE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2WJ9
 
 | | ArdB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Weikart, N.D, Roberts, G, Johnson, K.A, Oke, M, Cooper, L.P, McMahon, S.A, White, J.H, Liu, H, Carter, L.G, Walkinshaw, M.D, Blakely, G.W, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
3VQH
 
 | | Bromine SAD partially resolves multiple binding modes for PKA inhibitor H-89 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Pflug, A, Johnson, K.A, Engh, R.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anomalous dispersion analysis of inhibitor flexibility: a case study of the kinase inhibitor H-89
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1OGQ
 
 | | The crystal structure of PGIP (polygalacturonase inhibiting protein), a leucine rich repeat protein involved in plant defense | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, POLYGALACTURONASE INHIBITING PROTEIN | | Authors: | Di Matteo, A, Federici, L, Mattei, B, Salvi, G, Johnson, K.A, Savino, C, De Lorenzo, G, Tsernoglou, D, Cervone, F. | | Deposit date: | 2003-05-08 | | Release date: | 2003-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polygalacturonase-Inhibiting Protein (Pgip), a Leucine-Rich Repeat Protein Involved in Plant Defense
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
