1JEP
 
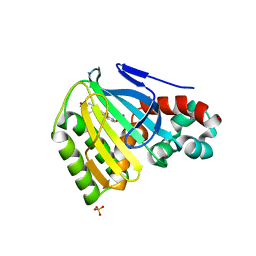 | | Chalcone Isomerase Complexed with 4'-hydroxyflavanone | | Descriptor: | 2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, CHALCONE--FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Noel, J.P. | | Deposit date: | 2001-06-18 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences.
J.Biol.Chem., 277, 2002
|
|
4K0N
 
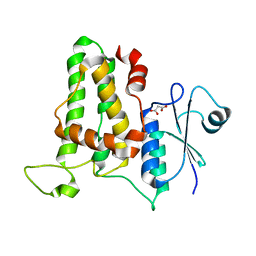 | | Crystal structure of human CLIC1 C24A mutant | | Descriptor: | Chloride intracellular channel protein 1, MALONATE ION | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure analysis of CLIC1 C24 mutants
To be Published
|
|
2BBO
 
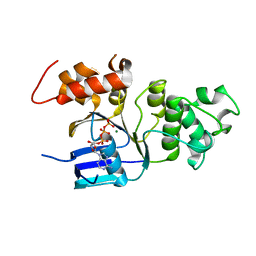 | | Human NBD1 with Phe508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
2AH4
 
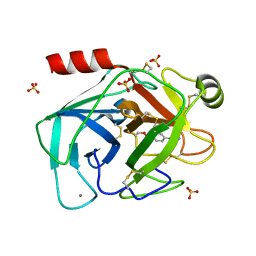 | | guanidinobenzoyl-trypsin acyl-enzyme at 1.13 A resolution | | Descriptor: | 4-carbamimidamidobenzoic acid, CALCIUM ION, SULFATE ION, ... | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AU4
 
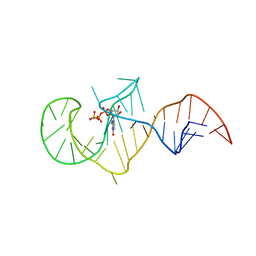 | | Class I GTP aptamer | | Descriptor: | Class I RNA aptamer to GTP, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Carothers, J.M, Davis, J.H, Chou, J.J, Szostak, J.W. | | Deposit date: | 2005-08-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an informationally complex high-affinity RNA aptamer to GTP.
Rna, 12, 2006
|
|
2W0U
 
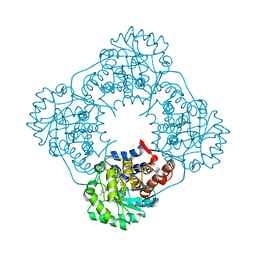 | | CRYSTAL STRUCTURE OF HUMAN GLYCOLATE OXIDASE IN COMPLEX WITH THE INHIBITOR 5-[(4-CHLOROPHENYL)SULFANYL]- 1,2,3-THIADIAZOLE-4-CARBOXYLATE. | | Descriptor: | 5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylate, FLAVIN MONONUCLEOTIDE, HYDROXYACID OXIDASE 1 | | Authors: | Bourhis, J.M, Lindqvist, Y. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Human Glycolate Oxidase in Complex with the Inhibitor 4-Carboxy-5-[(4-Chlorophenyl)Sulfanyl]-1,2,3-Thiadiazole.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2AVS
 
 | | kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
3QD4
 
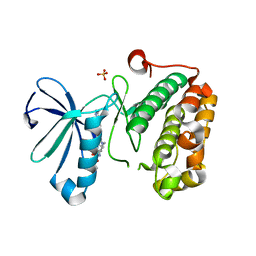 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl{(3R,5R)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-5-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, SULFATE ION, tert-butyl {(3R,5R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-5-methylpiperidin-3-yl}carbamate | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
2AXF
 
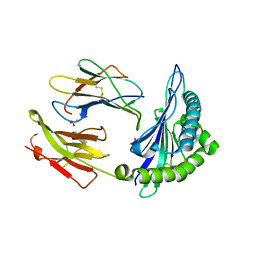 | | The Immunogenicity of a Viral Cytotoxic T Cell Epitope is controlled by its MHC-bound Conformation | | Descriptor: | 10-mer peptide from BZLF1 trans-activator protein, ACETIC ACID, Beta-2-microglobulin, ... | | Authors: | Tynan, F.E, Elhassen, D, Purcell, A.W, Burrows, J.M, Borg, N.A, Miles, J.J, Williamson, N.A, Green, K.J, Tellam, J, Kjer-Nielsen, L, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2005-09-05 | | Release date: | 2005-11-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation
J.Exp.Med., 202, 2005
|
|
2AM3
 
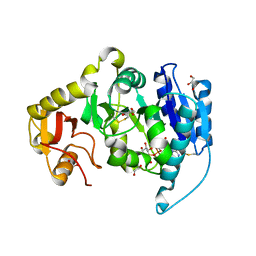 | |
2AZH
 
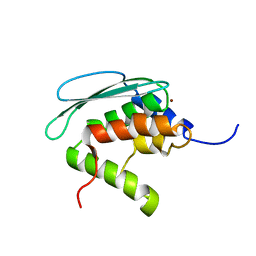 | | Solution structure of iron-sulfur cluster assembly protein SUFU from Bacillus subtilis, with zinc bound at the active site. Northeast Structural Genomics Consortium target SR17 | | Descriptor: | SufU, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Aramini, J.M, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-10 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Zn-Ligated Fe-S Cluster Assembly Scaffold Protein SufU From Bacillus subtilis
To be published
|
|
3TU6
 
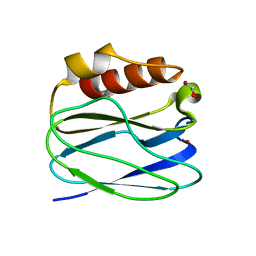 | | The Structure of a Pseudoazurin From Sinorhizobium meliltoi | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin (Blue copper protein) | | Authors: | Laming, E.M, McGrath, A.P, Guss, J.M, Maher, M.J. | | Deposit date: | 2011-09-16 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray crystal structure of a pseudoazurin from Sinorhizobium meliloti.
J.Inorg.Biochem., 115, 2012
|
|
3QD3
 
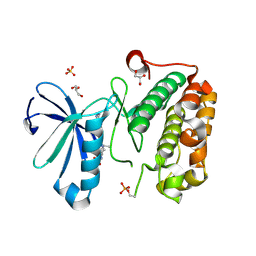 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl {(3R,6S)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
2VVE
 
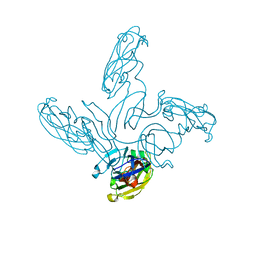 | | Crystal structure of the stem and receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2B4D
 
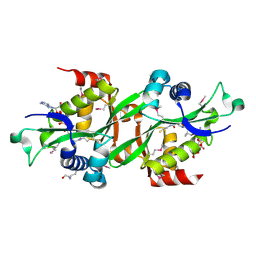 | | SSAT+COA+SP- SP disordered | | Descriptor: | COENZYME A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-23 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4K8I
 
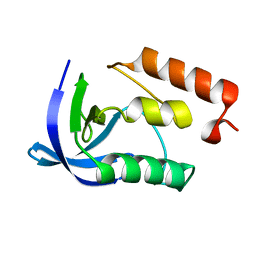 | | Crystal Structure of Staphylococcal Nuclease mutant I92V/V99L | | Descriptor: | Thermonuclease | | Authors: | Sanders, J.M, Latimer, E.C, Roeser, J.R, Janowska, K, Sakon, J, Stites, W.E. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic core mutants of Staphylococcal nuclease
To be Published
|
|
2VVF
 
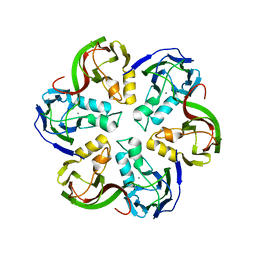 | | Crystal structure of the major capsid protein P2 from Bacteriophage PM2 | | Descriptor: | CALCIUM ION, MAJOR CAPSID PROTEIN P2 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2CK4
 
 | |
1FPT
 
 | |
2PX8
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and 7M-GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2PPG
 
 | | Crystal structure of putative isomerase from Sinorhizobium meliloti | | Descriptor: | Putative isomerase | | Authors: | Ramagopal, U.A, Toro, R, Dickey, M, Logan, C, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative isomerase from Sinorhizobium meliloti.
To be Published
|
|
4KSE
 
 | | Crystal structure of a HIV p51 (219-230) deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, HIV p51 subunit | | Authors: | Zheng, X, Mueller, G.A, Derose, E.F, Pedersen, L.C, Gabel, S.A, Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2013-05-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.
Nucleic Acids Res., 42, 2014
|
|
4LQR
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
3QCS
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-[2-Amino-6-(4-morpholinyl)-4-pyrimidinyl]-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-[2-amino-6-(morpholin-4-yl)pyrimidin-4-yl]-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
2PXC
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAM and GTPA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYLMETHIONINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
