2UYL
 
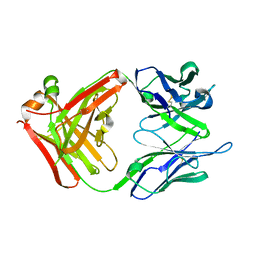 | | Crystal structure of a monoclonal antibody directed against an antigenic determinant common to Ogawa and Inaba serotypes of Vibrio cholerae O1 | | Descriptor: | MONOCLONAL ANTIBODY F-22-30 | | Authors: | Ahmed, F, Haouz, A, Nato, F, Fournier, J.M, Alzari, P.M. | | Deposit date: | 2007-04-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Monoclonal Antibody Directed Against an Antigenic Determinant Common to Ogawa and Inaba Serotypes of Vibrio Cholerae O1.
Proteins, 70, 2008
|
|
4KRG
 
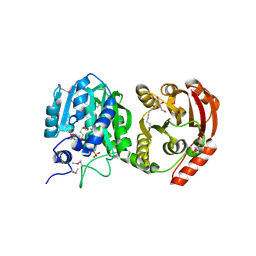 | |
2PMR
 
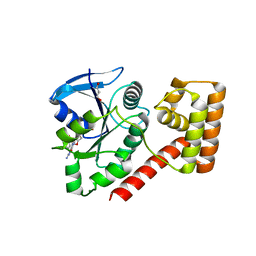
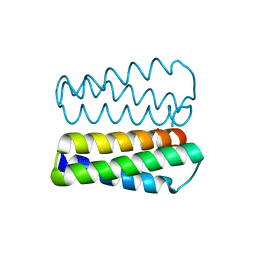 | | Crystal structure of a protein of unknown function from Methanobacterium thermoautotrophicum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structure of a protein of unknown function from Methanobacterium thermoautotrophicum.
To be Published
|
|
3TAH
 
 | | Crystal structure of an S. thermophilus NFeoB N11A mutant bound to mGDP | | Descriptor: | 3'-O-(N-methylanthraniloyl)guanosine-5'-diphosphate, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-08-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of an N11A mutant of the G-protein domain of FeoB
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2CK5
 
 | |
2PL7
 
 | |
4LN3
 
 | | The crystal structure of hemagglutinin from a H7N9 influenza virus (A/Shanghai/1/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Analysis of the Hemagglutinin from the Recent 2013 H7N9 Influenza Virus.
J.Virol., 87, 2013
|
|
4LN4
 
 | | The crystal structure of hemagglutinin form a h7n9 influenza virus (a/shanghai/1/2013) in complex with lstb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Analysis of the Hemagglutinin from the Recent 2013 H7N9 Influenza Virus.
J.Virol., 87, 2013
|
|
2PQL
 
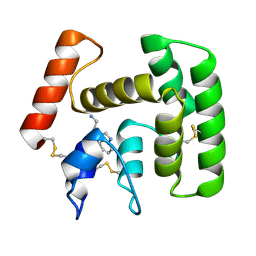 | | Crystal Structure of Anopheles gambiae D7R4-tryptamine complex | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, D7r4 protein | | Authors: | Andersen, J.F, Mans, B.J, Calvo, E, Ribeiro, J.M. | | Deposit date: | 2007-05-02 | | Release date: | 2007-10-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of D7r4, a Salivary Biogenic Amine-binding Protein from the Malaria Mosquito Anopheles gambiae.
J.Biol.Chem., 282, 2007
|
|
2PFQ
 
 | | Manganese promotes catalysis in a DNA polymerase lambda-DNA crystal | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, Downstream Primer, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
2PYF
 
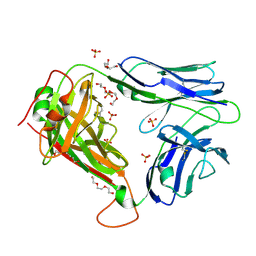 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry Unbound TCR Clone 5-1 | | Descriptor: | SULFATE ION, T-Cell Receptor, Alpha Chain, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
4LP7
 
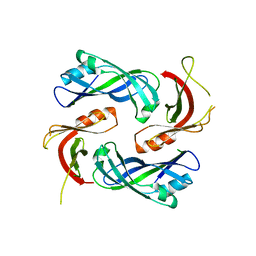 | |
1JE5
 
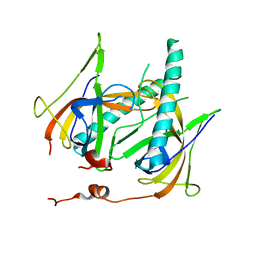 | | Crystal Structure of gp2.5, a Single-Stranded DNA Binding Protein Encoded by Bacteriophage T7 | | Descriptor: | CALCIUM ION, HELIX-DESTABILIZING PROTEIN | | Authors: | Hollis, T, Stattel, J.M, Walther, D.S, Richardson, C.C, Ellenberger, T.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-08-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2PMB
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Zhan, C, Shi, W, Toro, R, Sauder, J.M, Gilmore, J, Iizuka, M, Maletic, M, Gheyi, T, Wasserman, S.R, Smith, D, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Vibrio Cholerae.
To be Published
|
|
4M1Y
 
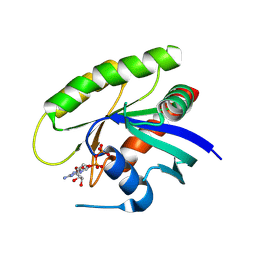 | | Crystal Structure of small molecule vinylsulfonamide 15 covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, N-{1-[N-(5,7-dichloro-2,1,3-benzothiadiazol-4-yl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
1CDB
 
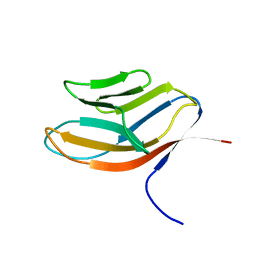 | |
1JUG
 
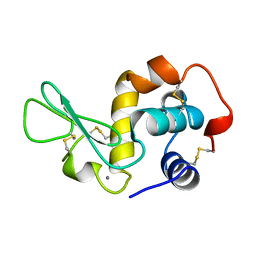 | | LYSOZYME FROM ECHIDNA MILK (TACHYGLOSSUS ACULEATUS) | | Descriptor: | CALCIUM ION, LYSOZYME | | Authors: | Guss, J.M. | | Deposit date: | 1996-10-13 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the calcium-binding echidna milk lysozyme at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4KM5
 
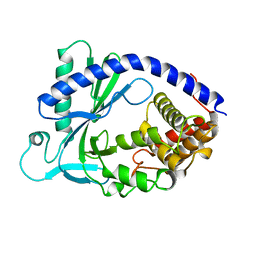 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
2CG6
 
 | | Second and third fibronectin type I module pair (crystal form I). | | Descriptor: | HUMAN FIBRONECTIN, SULFATE ION | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
4K8Y
 
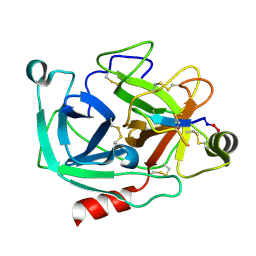 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with Sunflower Trypsin Inhibitor (SFTI-1) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
2V6H
 
 | | Crystal structure of the C1 domain of cardiac myosin binding protein-C | | Descriptor: | MYOSIN-BINDING PROTEIN C, CARDIAC-TYPE | | Authors: | Govata, L, Carpenter, L, Da Fonseca, P.C.A, Helliwell, J.R, Rizkallah, P.J, Flashman, E, Chayen, N.E, Redwood, C, Squire, J.M. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the C1 domain of cardiac myosin binding protein-C: implications for hypertrophic cardiomyopathy.
J. Mol. Biol., 378, 2008
|
|
1K1T
 
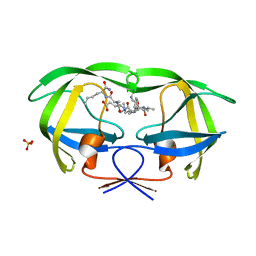 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN, SULFATE ION | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1BQ0
 
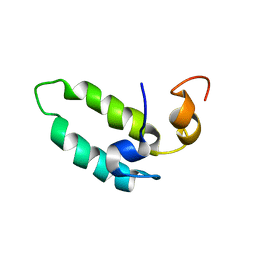 | |
1QOR
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI QUINONE OXIDOREDUCTASE COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, QUINONE OXIDOREDUCTASE, SULFATE ION | | Authors: | Thorn, J.M, Barton, J.D, Dixon, N.E, Ollis, D.L, Edwards, K.J. | | Deposit date: | 1995-02-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH.
J.Mol.Biol., 249, 1995
|
|
4KEL
 
 | | Atomic resolution crystal structure of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR(N) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
