4NQ4
 
 | |
4NFP
 
 | | Crystal Structure Analysis of the 16mer GCAGNCUUAAGUCUGC containing 8-aza-7-deaza-7-ethynyl Adenosine | | Descriptor: | FORMAMIDE, GCAG(A7E)CUUAAGUCUGC | | Authors: | Beal, P.A, Fisher, A.J, Phelps, K.J, Ibarra-Soza, J.M, Zheng, Y. | | Deposit date: | 2013-10-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Click Modification of RNA at Adenosine: Structure and Reactivity of 7-Ethynyl- and 7-Triazolyl-8-aza-7-deazaadenosine in RNA.
Acs Chem.Biol., 9, 2014
|
|
4NFQ
 
 | | Crystal Structure Analysis of the 16mer GCAGNCUUAAGUCUGC containing 7-triazolyl-8-aza-7-deazaadenosine | | Descriptor: | GCAG(7AT)CUUAAGUCUGC | | Authors: | Beal, P.A, Fisher, A.J, Phelps, K.J, Ibarra-Soza, J.M, Zheng, Y. | | Deposit date: | 2013-10-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Click Modification of RNA at Adenosine: Structure and Reactivity of 7-Ethynyl- and 7-Triazolyl-8-aza-7-deazaadenosine in RNA.
Acs Chem.Biol., 9, 2014
|
|
1P6A
 
 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
4QNH
 
 | | Calcium-calmodulin (T79D) complexed with the calmodulin binding domain from a small conductance potassium channel SK2-a | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Zhang, M, Pascal, J.M, Logothetis, D.E, Zhang, J.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selective phosphorylation modulates the PIP2 sensitivity of the CaM-SK channel complex.
Nat.Chem.Biol., 10, 2014
|
|
1G10
 
 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
4MX1
 
 | | Structure of ricin A chain bound with 2-amino-4-oxo-N-(2-(3-phenylureido)ethyl)-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-amino-4-oxo-N-{2-[(phenylcarbamoyl)amino]ethyl}-3,4-dihydropteridine-7-carboxamide, MALONIC ACID, Ricin A chain, ... | | Authors: | Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Jasheway, K.R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4N4A
 
 | | Cystal structure of Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
4QQV
 
 | | Extracellular domains of mouse IL-3 beta receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-3 receptor class 2 subunit beta | | Authors: | Jackson, C.J, Young, I.G, Murphy, J.M, Carr, P.D, Ewens, C.L, Dai, J, Ollis, D.L. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the mouse interleukin-3 beta-receptor: insights into interleukin-3 binding and receptor activation.
Biochem.J., 463, 2014
|
|
4QYY
 
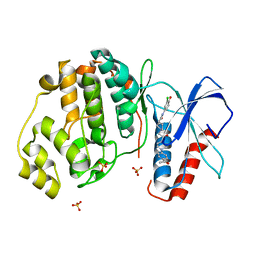 | | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase State | | Descriptor: | (3R)-1-{2-[4-(4-acetylphenyl)piperazin-1-yl]-2-oxoethyl}-N-(3-chloro-4-hydroxyphenyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Deng, Y, Shipps, G.W, Cooper, A, English, J.M, Annis, D.A, Carr, D, Nan, Y, Wang, T, Zhu, Y.H, Chuang, C, Dayananth, P, Hruza, A.W, Xiao, L, Jin, W, Kirschmeier, P, Windsor, W.T, Samatar, A.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase.
J.Med.Chem., 57, 2014
|
|
4R6W
 
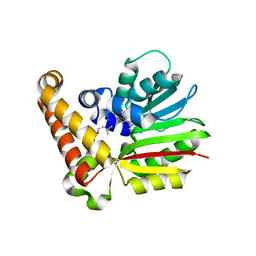 | |
3IWZ
 
 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
4R1Q
 
 | |
4R2Y
 
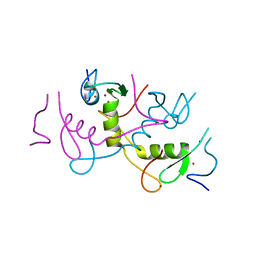 | | Crystal structure of APC11 RING domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissmann, F, Jarvis, M.A, Vanderlinden, R, Grace, C.R.R, Frye, J.J, Dube, P, Qiao, R, Petzold, G, Cho, S.E, Alsharif, O, Bao, J, Zheng, J, Nourse, A, Kurinov, I, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
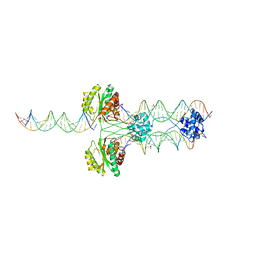
4R79
 
 | |
4R0E
 
 | |
4R0R
 
 | | Ebolavirus GP Prehairpin Intermediate Mimic | | Descriptor: | eboIZN21 | | Authors: | Clinton, T.R, Weinstock, M.T, Jacobsen, M.T, Szabo-Fresnais, N, Pandya, M.J, Whitby, F.G, Herbert, A.S, Prugar, L.I, McKinnon, R, Hill, C.P, Welch, B.D, Dye, J.M, Eckert, D.M, Kay, M.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and characterization of ebolavirus GP prehairpin intermediate mimics as drug targets.
Protein Sci., 24, 2015
|
|
4R1O
 
 | |
4R27
 
 | | Crystal structure of beta-glycosidase BGL167 | | Descriptor: | Glycoside hydrolase | | Authors: | Park, S.J, Choi, J.M, Kyeong, H.H, Kim, S.G, Kim, H.S. | | Deposit date: | 2014-08-09 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational design of a beta-glycosidase with high regiospecificity for triterpenoid tailoring
Chembiochem, 16, 2015
|
|
1K38
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2 | | Descriptor: | Beta-lactamase OXA-2, FORMIC ACID | | Authors: | Kerff, F, Fonze, E, Bouillenne, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
4E2K
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain | | Descriptor: | BENZAMIDINE, DNA primase | | Authors: | Rymer, R.U, Solorio, F.A, Clement, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-08 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
8PUJ
 
 | | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin. | | Descriptor: | COB(II)INAMIDE, HYDROXIDE ION, SODIUM ION, ... | | Authors: | Whittaker, J, Martinez-Felices, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The surface-exposed lipo-protein of BtuG3 in complex with cyanocobinamide.
To Be Published
|
|
8PTF
 
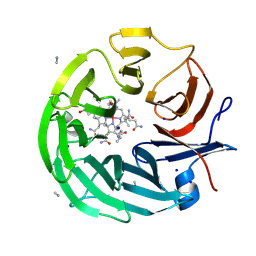 | | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin. | | Descriptor: | COB(II)INAMIDE, SODIUM ION, YncE family protein, ... | | Authors: | Whittaker, J, Felices Martinez, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2023-07-14 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The surface-exposed lipo-protein of BtuG1 in complex with cyanocobinamide.
To Be Published
|
|
1AR6
 
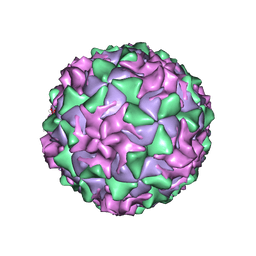 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
4EEU
 
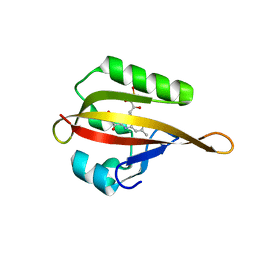 | | Crystal structure of phiLOV2.1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4068 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
