7EKB
 
 | | Crystal structure of 4E10 modified with pyrene acetamide | | Descriptor: | ACETATE ION, Fab region of the heavy chain of broadly neutralizing antibody anti-HIV-1 4E10, Fab region of the light chain of the broadly neutralizing anti-HIV-1 antibody 4E10, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
6P9E
 
 | | Crystal structure of IL-36gamma complexed to A-552 | | Descriptor: | (2S)-2-{[4-(3-amino-4-methylphenyl)-6-methylpyrimidin-2-yl]oxy}-3-methoxy-3,3-diphenylpropanoic acid, Interleukin-36 gamma | | Authors: | Argiriadi, M.A, Todorovic, T, Su, Z, Putman, B, Kakavas, S.J, Salte, K.M, McDonald, H.A, Wetter, J.B, Paulsboe, S.E, Sun, Q, Gerstein, C.E, Medina, L, Sielaff, B, Sadhukhan, R, Stockmann, H, Richardson, P.L, Qiu, W, Henry, R.F, Herold, J.M, Shotwell, J.B, McGaraughty, S.P, Honore, P, Gopalakrishnan, S.M, Sun, C.C, Scott, V.E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecule IL-36 gamma Antagonist as a Novel Therapeutic Approach for Plaque Psoriasis.
Sci Rep, 9, 2019
|
|
7K9H
 
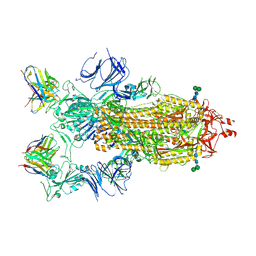 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
7EKK
 
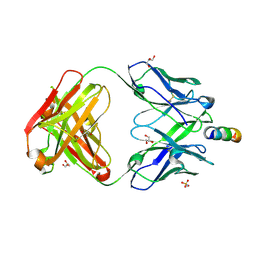 | | Anti-HIV-1 broadly neutralizing antibody delta-loop 4E10 modified with pyrene acetamide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
7K9I
 
 | |
4XKT
 
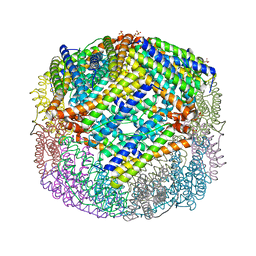 | | E coli BFR variant Y149F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Bradley, J.M, Hemmings, A.M, Le Brun, N.E. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6PDT
 
 | | cryoEM structure of yeast glucokinase filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glucokinase-1, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dosey, A.M, Farrell, D.P, Stoddard, P.R, Kollman, J.M. | | Deposit date: | 2019-06-19 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Polymerization in the actin ATPase clan regulates hexokinase activity in yeast.
Science, 367, 2020
|
|
7K9J
 
 | |
5QD2
 
 | | Crystal structure of BACE complex with BMC017 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(methoxymethyl)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDC
 
 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | Descriptor: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6P7D
 
 | | D104N S. typhimurium siroheme synthase | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
7KDQ
 
 | |
6P90
 
 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
7Q0L
 
 | | Crystal structure of the peptide transporter YePEPT-K314A at 2.93 A | | Descriptor: | Peptide transporter YePEPT | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Boggavarapu, R, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
5QDA
 
 | | Crystal structure of BACE complex with BMC013 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-18-methoxy-3,15,17-triazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
7Q0M
 
 | | Crystal structure of the peptide transporter YePEPT-K314A in complex with LZNV at 2.66 A | | Descriptor: | (2~{S})-2-[[(2~{S})-2-azanyl-6-[(4-nitrophenyl)methoxycarbonylamino]hexanoyl]amino]-3-methyl-butanoic acid, Peptide transporter YePEPT, UNDECYL-MALTOSIDE | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
6PDN
 
 | | Human PIM1 bound to benzothiophene inhibitor 292 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxylic acid, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
6PDO
 
 | | Human PIM1 bound to benzothiophene inhibitor 354 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxamide, Peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
4XKU
 
 | | E coli BFR variant Y114F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Hemmings, A.M, Le Brun, N.E, Bradley, J.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5QCO
 
 | | Crystal structure of BACE complex with BMC016 | | Descriptor: | (4S)-19-acetyl-4-[(1R)-1-hydroxy-2-({1-[3-(propan-2-yl)phenyl]cyclopropyl}amino)ethyl]-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDD
 
 | | Crystal structure of BACE complex with BMC020 hydrolyzed | | Descriptor: | (10R,12S)-12-[(1R)-1,2-dihydroxyethyl]-N,N,10-trimethyl-14-oxo-2-oxa-13-azabicyclo[13.3.1]nonadeca-1(19),15,17-triene-17-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6PHS
 
 | | Protein Tyrosine Phosphatase 1B (1-301), P185A mutant, vanadate bound state | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-06-25 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
7KVB
 
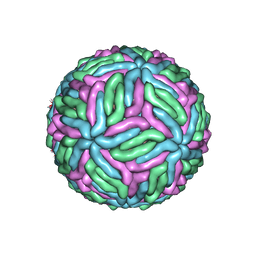 | | Chimeric flavivirus between Binjari virus and Murray Valley encephalitis virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
4XUC
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd18 (1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one) | | Descriptor: | 1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
6PR2
 
 | | R261A/S128A S. typhimurium siroheme synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
