1JNF
 
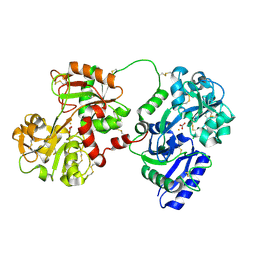 | | Rabbit serum transferrin at 2.6 A resolution. | | Descriptor: | CARBONATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-24 | | Release date: | 2001-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 A, respectively.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2VVW
 
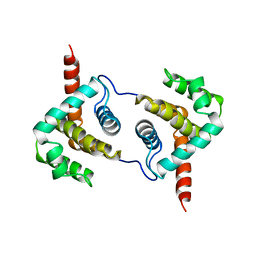 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2CME
 
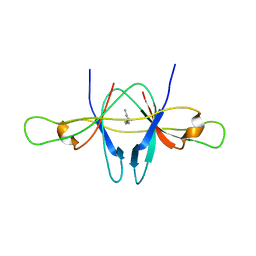 | | The crystal structure of SARS coronavirus ORF-9b protein | | Descriptor: | DECANE, HYPOTHETICAL PROTEIN 5 | | Authors: | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2006-05-06 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
2VSM
 
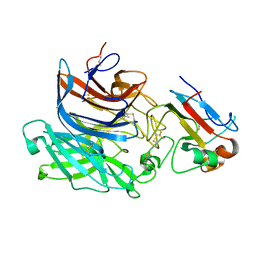 | | Nipah virus attachment glycoprotein in complex with human cell surface receptor ephrinB2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EPHRIN-B2, HEMAGGLUTININ-NEURAMINIDASE, ... | | Authors: | Bowden, T.A, Aricescu, A.R, Gilbert, R.J, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Nipah and Hendra Virus Attachment to Their Cell-Surface Receptor Ephrin-B2
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VVD
 
 | | Crystal structure of the receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2W2R
 
 | | Structure of the vesicular stomatitis virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2WBG
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, ACETATE ION, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
2W0I
 
 | | Structure Of C-Terminal Actin Depolymerizing Factor Homology (Adf-H) Domain Of Human Twinfilin-2 | | Descriptor: | TWINFILIN-2 | | Authors: | Elkins, J.M, Pike, A.C.W, Salah, E, Savitsky, P, Murray, J.W, Roos, A, von Delft, F, Edwards, A, Arrowsmith, C.H, Wikstrom, M, Bountra, C, Knapp, S. | | Deposit date: | 2008-08-18 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of C-Terminal Actin Depolymerizing Factor Homology (Adf-H) Domain of Human Twinfilin- 2
To be Published
|
|
3QCQ
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-(3-Amino-1H-indazol-6-yl)-N4-ethyl-2,4-pyrimidinediamine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-(3-amino-2H-indazol-6-yl)-N~4~-ethylpyrimidine-2,4-diamine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
2A1L
 
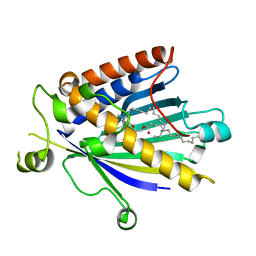 | | Rat PITP-Beta Complexed to Phosphatidylcholine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylinositol transfer protein beta isoform | | Authors: | Vordtriede, P.B, Doan, C.N, Tremblay, J.M, Helmkamp, G.M, Yoder, M.D. | | Deposit date: | 2005-06-20 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure of PITPbeta in Complex with Phosphatidylcholine: Comparison of Structure and Lipid Transfer to Other PITP Isoforms.
Biochemistry, 44, 2005
|
|
2VQP
 
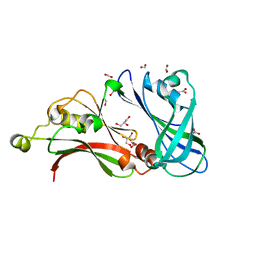 | | Structure of the matrix protein from human Respiratory Syncytial Virus | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Money, V.A, McPhee, H.K, Sanderson, J.M, Yeo, R.P. | | Deposit date: | 2008-03-18 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Surface Features of a Mononegavirales Matrix Protein Indicate Sites of Membrane Interaction.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3U3L
 
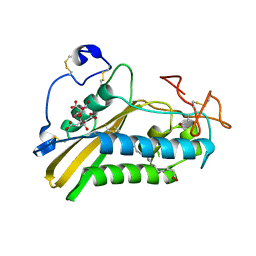 | | Crystal structure of the selenomethionine derivative of tablysin-15 | | Descriptor: | CITRIC ACID, PALMITIC ACID, PRASEODYMIUM ION, ... | | Authors: | Andersen, J.F, Xu, X, Ribeiro, J.M. | | Deposit date: | 2011-10-06 | | Release date: | 2012-02-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain.
J.Biol.Chem., 287, 2012
|
|
2VWR
 
 | | Crystal structure of the second pdz domain of numb-binding protein 2 | | Descriptor: | LIGAND OF NUMB PROTEIN X 2 | | Authors: | Roos, A.K, Guo, K, Burgess-Brown, N, Yue, W.W, Elkins, J.M, Pike, A.C.W, Filippakopoulos, P, Arrowsmith, C.H, Wikstom, M, Edwards, A, von Delft, F, Bountra, C, Doyle, D, Oppermann, U. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the Second Pdz Domain of the Human Numb-Binding Protein 2
To be Published
|
|
4IBP
 
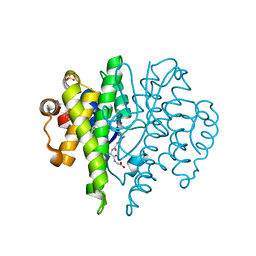 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-like protein YibF, SULFATE ION | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target IFI-900011, with bound glutathione
To be Published
|
|
2VVY
 
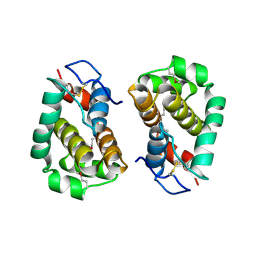 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2VWD
 
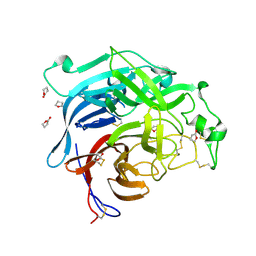 | | Nipah Virus Attachment Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-BUTYROLACTONE, ... | | Authors: | Bowden, T.A, Crispin, M, Harvey, D.J, Aricescu, A.R, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure and Carbohydrate Analysis of Nipah Virus Attachment Glycoprotein: A Template for Antiviral and Vaccine Design.
J.Virol., 82, 2008
|
|
4ICN
 
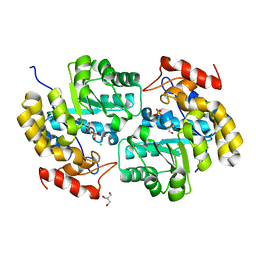 | | Dihydrodipicolinate synthase from shewanella benthica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDRODIPICOLINATE SYNTHASE, ... | | Authors: | Wubben, J.M, Paxman, J.J, Dogovski, C, Parker, M.W, Perugini, M.A. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cold enzymology offers insight into molecular evolution in quaternary structure
To be Published
|
|
4ID0
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione sulfinic acid (gso2h) and acetate | | Descriptor: | ACETATE ION, GLYCEROL, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-11 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione sulfinic acid (gso2h) and acetate
To be Published
|
|
2W2F
 
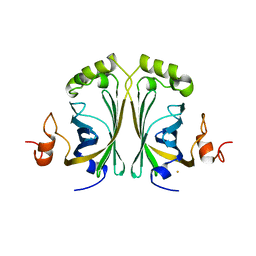 | | CRYSTAL STRUCTURE OF SINGLE POINT MUTANT ARG48GLN OF P-COUMARIC ACID DECARBOXYLASE FROM LACTOBACILLUS PLANTARUM STRUCTURAL INSIGHTS INTO THE ACTIVE SITE AND DECARBOXYLATION CATALYTIC MECHANISM | | Descriptor: | BARIUM ION, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-29 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
1IDC
 
 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
3TLJ
 
 | | Crystal structure of Trm14 from Pyrococcus furiosus in complex with S-adenosyl-L-homocysteine | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine N2-)-methyltransferase Trm14 | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-30 | | Release date: | 2012-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
2RD7
 
 | | Human Complement Membrane Attack Proteins Share a Common Fold with Bacterial Cytolysins | | Descriptor: | CHLORIDE ION, Complement component C8 alpha chain, Complement component C8 gamma chain | | Authors: | Slade, D.J, Lovelace, L.L, Chruszcz, M, Minor, W, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-09-21 | | Release date: | 2008-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.
J. Biol. Chem., 286, 2011
|
|
3TM5
 
 | | Crystal structure of Trm14 from Pyrococcus furiosus in complex with sinefungin | | Descriptor: | Crystal structure of Trm14, SINEFUNGIN | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-31 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
4LPK
 
 | | Crystal Structure of K-Ras WT, GDP-bound | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
1IDF
 
 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
