8BAI
 
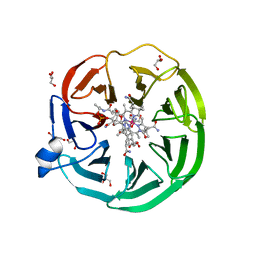 | | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin. | | Descriptor: | CYANOCOBALAMIN, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J, Martinez Felices, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin.
To Be Published
|
|
8BB0
 
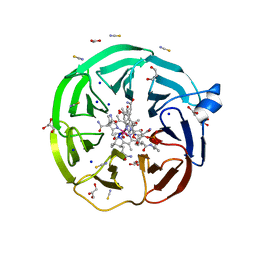 | | The surface-exposed lipo-protein of BtuG2 in complex with hydroxycobalamin. | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J, Felices Martinez, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The surface-exposed lipo-protein of BtuG2 in complex with hydroxycobinamide.
To Be Published
|
|
2ND3
 
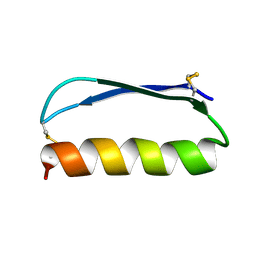 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
1G2G
 
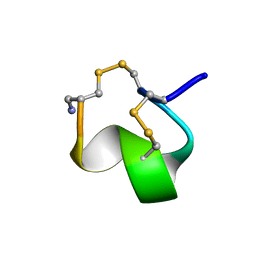 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
1PIV
 
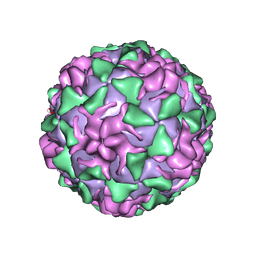 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1JBP
 
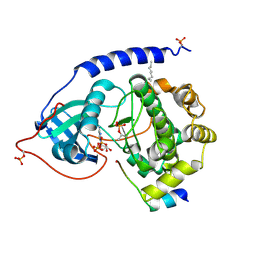 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Substrate Peptide, ADP and Detergent | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, MUSCLE/BRAIN FORM, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
5DEK
 
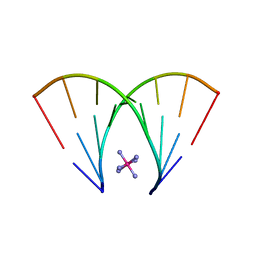 | | RNA octamer containing dT | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing dT | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
4OI9
 
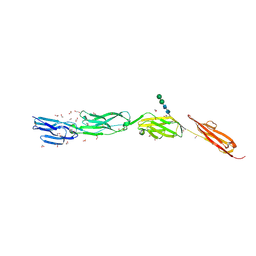 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NMN
 
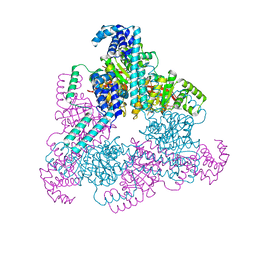 | | Aquifex aeolicus replicative helicase (DnaB) complexed with ADP, at 3.3 resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Lyubimov, A.Y, Strycharska, M.S, Erzberger, J.P, Berger, J.M. | | Deposit date: | 2013-11-15 | | Release date: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Nucleotide and partner-protein control of bacterial replicative helicase structure and function.
Mol.Cell, 52, 2013
|
|
4NQ5
 
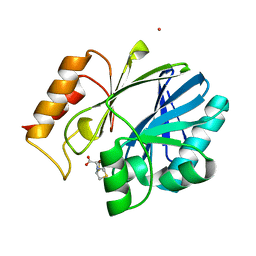 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Inhibition of metallo-lactamases with bicyclic compounds
TO BE PUBLISHED
|
|
4NDZ
 
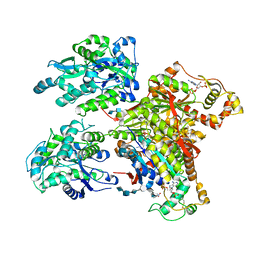 | | Structure of Maltose Binding Protein fusion to 2-O-Sulfotransferase with bound heptasaccharide and PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1 fusion, ... | | Authors: | Liu, C, Sheng, J, Krahn, J.M, Perera, L, Xu, Y, Hsieh, P, Liu, J, Pedersen, L.C. | | Deposit date: | 2013-10-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Deciphering the role of 2-O-sulfotransferase in regulating heparan sulfate biosynthesis
To be Published
|
|
4PXT
 
 | |
3VEU
 
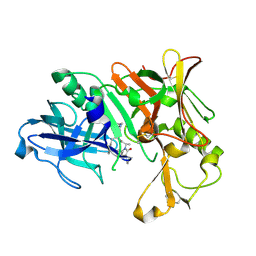 | | Crystal Structure of Human Beta Secretase in Complex with NVP-AVI326 | | Descriptor: | (2S)-N-[(2S,3R)-3-hydroxy-1-phenyl-4-{[3-(propan-2-yl)benzyl]amino}butan-2-yl]-2-[(5S)-6-oxo-1-propyl-1,7-diazaspiro[4.4]non-7-yl]propanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
4P0D
 
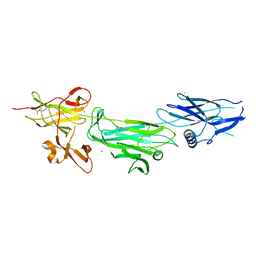 | | The T6 backbone pilin of serotype M6 Streptococcus pyogenes has a modular three-domain structure decorated with variable loops and extensions | | Descriptor: | CALCIUM ION, IODIDE ION, Trypsin-resistant surface T6 protein | | Authors: | Young, P.G, Moreland, N.J, Loh, J.M, Bell, A, Atatoa-Carr, P, Proft, T, Baker, E.N. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Conservation, Variability, and Immunogenicity of the T6 Backbone Pilin of Serotype M6 Streptococcus pyogenes.
Infect.Immun., 82, 2014
|
|
1M3V
 
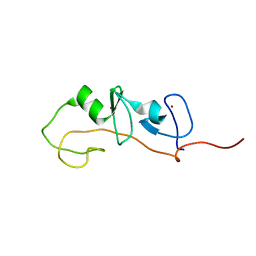 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1MBT
 
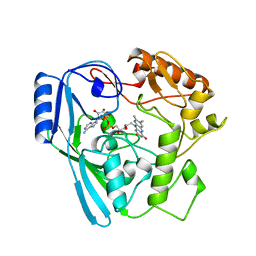 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls.
Structure, 4, 1996
|
|
1MBB
 
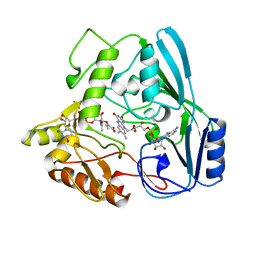 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-3(N-ACETYLGLUCOSAMINYL)BUTYRIC ACID | | Authors: | Benson, T.E, Lees, W.J, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-07 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | (E)-enolbutyryl-UDP-N-acetylglucosamine as a mechanistic probe of UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
Biochemistry, 35, 1996
|
|
1JLU
 
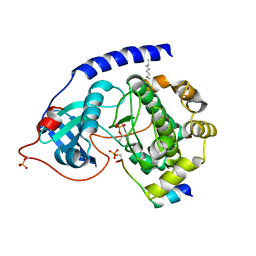 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
4PPV
 
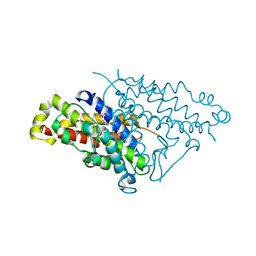 | |
4PED
 
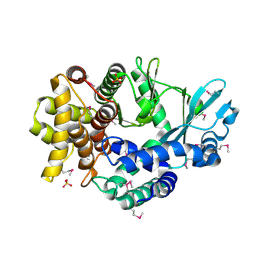 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
4PQH
 
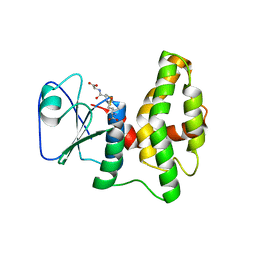 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
1PLC
 
 | |
4NDB
 
 | | X-ray structure of a mutant (T61D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1ONY
 
 | | Oxalyl-Aryl-Amino Benzoic Acid inhibitors of PTP1B, compound 17 | | Descriptor: | 2-{[2-(2-CARBAMOYL-VINYL)-4-(2-METHANESULFONYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-PHENYL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
4NDC
 
 | | X-ray structure of a mutant (T188D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
