2WFS
 
 | | Fitting of influenza virus NP structure into the 9-fold symmetryzed cryoEM reconstruction of an active RNP particle. | | Descriptor: | NUCLEOPROTEIN | | Authors: | Coloma, R, Valpuesta, J.M, Arranz, R, Carrascosa, J.L, Ortin, J, Martin-Benito, J. | | Deposit date: | 2009-04-15 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The Structure of a Biologically Active Influenza Virus Ribonucleoprotein Complex.
Plos Pathog., 5, 2009
|
|
1FVV
 
 | | THE STRUCTURE OF CDK2/CYCLIN A IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(7-OXO-7H-THIAZOLO[5,4-E]INDOL-8-YLMETHYL)-AMINO]-N-PYRIDIN-2-YL-BENZENESULFONAMIDE, CYCLIN A, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1ZH0
 
 | | Crystal Structure of L-3-(2-napthyl)alanine-tRNA synthetase in complex with L-3-(2-napthyl)alanine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-(2-NAPHTHYL)-ALANINE, Tyrosyl-tRNA synthetase | | Authors: | Turner, J.M, Graziano, J, Spraggon, G, Schultz, P.G. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of an aminoacyl-tRNA synthetase active site
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1G25
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE HUMAN TFIIH MAT1 SUBUNIT | | Descriptor: | CDK-ACTIVATING KINASE ASSEMBLY FACTOR MAT1, ZINC ION | | Authors: | Gervais, V, Wasielewski, E, Busso, D, Poterszman, A, Egly, J.M, Thierry, J.C, Kieffer, B. | | Deposit date: | 2000-10-17 | | Release date: | 2000-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Domain of the Human TFIIH MAT1 Subunit: New Insights into the RING Finger Family
J.Biol.Chem., 276, 2001
|
|
4LYJ
 
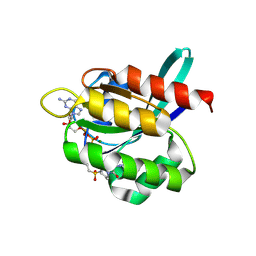 | | Crystal Structure of small molecule vinylsulfonamide 9 covalently bound to K-Ras G12C, alternative space group | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-31 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4MDA
 
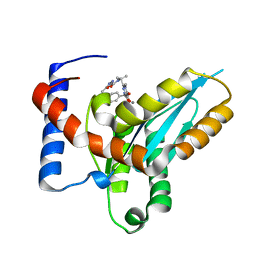 | | Structure of Mos1 transposase catalytic domain and Raltegravir with Mn | | Descriptor: | MANGANESE (II) ION, Mariner Mos1 transposase, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide | | Authors: | Richardson, J.M. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Mos1 Transposase Inhibition by the Anti-retroviral Drug Raltegravir.
Acs Chem.Biol., 9, 2014
|
|
4LZG
 
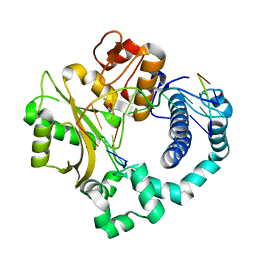 | | Binary complex of human DNA Polymerase Mu with DNA | | Descriptor: | CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, GLYCEROL, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2W4F
 
 | | CRYSTAL STRUCTURE OF THE FIRST PDZ DOMAIN OF HUMAN SCRIB1 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN LAP4 | | Authors: | Hozjan, V, Pilka, E.S, Roos, A.K, W Yue, W, Phillips, C, Bray, J, Cooper, C, Salah, E, Elkins, J.M, Muniz, J.R.C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Bountra, C, Doyle, D.A, Oppermann, U. | | Deposit date: | 2008-11-25 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the First Pdz Domain of Human Scrib1
To be Published
|
|
1J4X
 
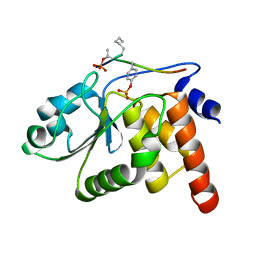 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE C124S MUTANT-PEPTIDE COMPLEX | | Descriptor: | DDE(AHP)(TPO)G(PTR)VATR, DUAL SPECIFICITY PROTEIN PHOSPHATASE 3 | | Authors: | Schumacher, M.A, Todd, J.L, Tanner, K.G, Denu, J.M. | | Deposit date: | 2001-12-13 | | Release date: | 2001-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein Phosphatase.
Biochemistry, 41, 2002
|
|
1YZS
 
 | |
1Z1C
 
 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute virus of Mice | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-D(*AP*CP*AP*CP*CP*AP*AP*AP*A)-3', 5'-D(*AP*TP*CP*CP*TP*CP*TP*AP*TP*CP*AP*C)-3', ... | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
2W37
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC CATABOLIC ORNITHINE TRANSCARBAMYLASE FROM Lactobacillus hilgardii | | Descriptor: | NICKEL (II) ION, ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | de las Rivas, B, Fox, G.C, Angulo, I, Rodriguez, H, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hexameric Catabolic Ornithine Transcarbamylase from Lactobacillus Hilgardii: Structural Insights Into the Oligomeric Assembly and Metal Binding.
J.Mol.Biol., 393, 2009
|
|
1Z5A
 
 | | Topoisomerase VI-B, ADP-bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
4IJ2
 
 | | Human methemoglobin in complex with the second and third NEAT domains of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein H, ... | | Authors: | Dickson, C.F, Jacques, D.A, Guss, J.M, Gell, D.A. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.24 Å) | | Cite: | Structure of the Hemoglobin-IsdH Complex Reveals the Molecular Basis of Iron Capture by Staphylococcus aureus
J.Biol.Chem., 289, 2014
|
|
1ZAW
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form A | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
2VWL
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3R,5S)-1-{[2-FLUORO-4-(2-OXO-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-5-HYDROXYMETHYL-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2W2A
 
 | | Crystal Structure of p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
1ZZY
 
 | |
1IRO
 
 | | RUBREDOXIN (OXIDIZED, FE(III)) AT 1.1 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M, Meyer, J. | | Deposit date: | 1995-12-13 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2WC3
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-8-epi-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8S,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
2A11
 
 | |
1TEY
 
 | | NMR structure of human histone chaperone, ASF1A | | Descriptor: | ASF1 anti-silencing function 1 homolog A | | Authors: | Mousson, F, Lautrette, A, Thuret, J.Y, Agez, M, Amigues, B, Courbeyrette, R, Neumann, J.M, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2004-05-26 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of Asf1 with histone H3 and its functional implications.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2VSK
 
 | | Hendra virus attachment glycoprotein in complex with human cell surface receptor ephrinB2 | | Descriptor: | EPHRIN-B2, HEMAGGLUTININ-NEURAMINIDASE | | Authors: | Bowden, T.A, Aricescu, A.R, Gilbert, R.J, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Nipah and Hendra Virus Attachment to Their Cell-Surface Receptor Ephrin-B2
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VVV
 
 | | Aminopyrrolidine-related triazole Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-1H-1,2,4-triazol-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
295D
 
 | | CRYSTAL AND SOLUTION STRUCTURES OF THE OLIGONUCLEOTIDE D(ATGCGCAT)2: A COMBINED X-RAY AND NMR STUDY | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*CP*AP*T)-3') | | Authors: | Clark, G.R, Brown, D.G, Sanderson, M.R, Chwalinski, T, Neidle, S, Veal, J.M, Jones, R.L, Wilson, W.D, Zon, G, Garman, E, Stuart, D.I. | | Deposit date: | 1991-05-28 | | Release date: | 1996-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal and solution structures of the oligonucleotide d(ATGCGCAT)2: a combined X-ray and NMR study.
Nucleic Acids Res., 18, 1990
|
|
