2BWY
 
 | | Glu383Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
6OQO
 
 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3W0L
 
 | | The crystal structure of Xenopus Glucokinase and Glucokinase Regulatory Protein complex | | Descriptor: | FRUCTOSE -6-PHOSPHATE, Glucokinase, Glucokinase regulatory protein, ... | | Authors: | Choi, J.M, Seo, M.H, Kyeong, H.H, Kim, E, Kim, H.S. | | Deposit date: | 2012-10-31 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6OQL
 
 | | CDK6 in complex with Cpd13 (R)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 6 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2V11
 
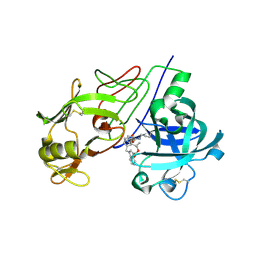 | | Crystal Structure of Renin with Inhibitor 6 | | Descriptor: | (2S,4S,5R,7R)-4-AMINO-8-(BUTYLAMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXOOCTYL 1-BENZYL-1H-INDOLE-3-CARBOXYLATE, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
3V90
 
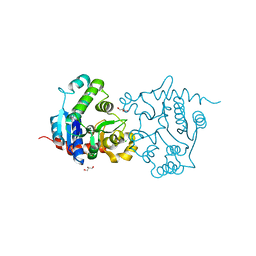 | | Structure of T82M glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
1ZG2
 
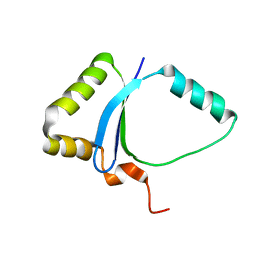 | | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2. | | Descriptor: | Hypothetical UPF0213 protein BH0048 | | Authors: | Aramini, J.M, Swapna, G.V.T, Xiao, R, Ma, L, Shastry, R, Ciano, M, Acton, T.B, Liu, J, Rost, B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2.
To be Published
|
|
3SKY
 
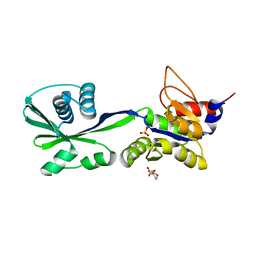 | | 2.1A crystal structure of the phosphate bound ATP binding domain of Archaeoglobus fulgidus COPB | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Copper-exporting P-type ATPase B, PHOSPHATE ION | | Authors: | Jayakanthan, S, Roberts, S.A, Weichsel, A, Arguello, J.M, McEvoy, M.M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformations of the apo-, substrate-bound and phosphate-bound ATP-binding domain of the Cu(II) ATPase CopB illustrate coupling of domain movement to the catalytic cycle.
Biosci.Rep., 32, 2012
|
|
1ZPH
 
 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8315 complexed with CGCGAATTCGCG | | Descriptor: | 1,6-DIMETHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3S95
 
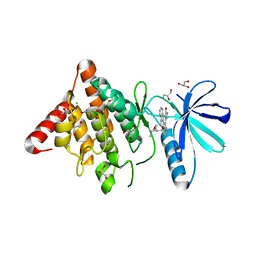 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
3SA1
 
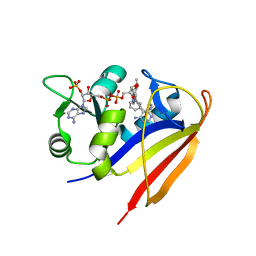 | |
1GUW
 
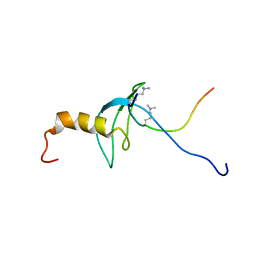 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
1Z2Q
 
 | |
2VGA
 
 | | The structure of Vaccinia virus A41 | | Descriptor: | PROTEIN A41 | | Authors: | Bahar, M.W, Kenyon, J.C, Putz, M.M, Abrescia, N.G.A, Pease, J.E, Wise, E.L, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function of A41, a Vaccinia Virus Chemokine Binding Protein.
Plos Pathog., 4, 2008
|
|
2VHT
 
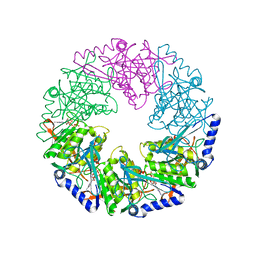 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 R279A mutant in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VKP
 
 | | Crystal structure of BTB domain from BTBD6 | | Descriptor: | 1,2-ETHANEDIOL, BTB/POZ DOMAIN-CONTAINING PROTEIN 6, POTASSIUM ION | | Authors: | Cooper, C.D.O, Pike, A.C.W, Salah, E, Filippakopoulos, P, Bunkoczi, G, Elkins, J.M, von Delft, F, Gileadi, O, Edwards, A, Weigelt, J, Arrowsmith, C.H, Knapp, S. | | Deposit date: | 2007-12-21 | | Release date: | 2008-02-12 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Btb Domain from Btbd6
To be Published
|
|
2V10
 
 | | Crystal Structure of Renin with Inhibitor 9 | | Descriptor: | (2R,4S,5S,7S)-5-AMINO-N-BUTYL-4-HYDROXY-7-[4-METHOXY-3-(3-METHOXYPROPOXY)BENZYL]-2,8-DIMETHYLNONANAMIDE, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
4L01
 
 | |
1ZUN
 
 | | Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Mougous, J.D, Lee, D.H, Hubbard, S.C, Schelle, M.W, Vocadlo, D.J, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for g protein control of the prokaryotic ATP sulfurylase.
Mol.Cell, 21, 2006
|
|
3G9Y
 
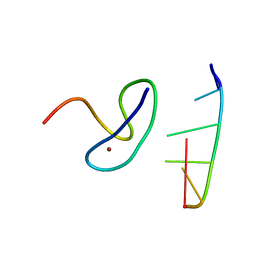 | | Crystal structure of the second zinc finger from ZRANB2/ZNF265 bound to 6 nt ssRNA sequence AGGUAA | | Descriptor: | RNA (5'-R(*AP*GP*GP*UP*AP*A)-3'), ZINC ION, Zinc finger Ran-binding domain-containing protein 2 | | Authors: | Loughlin, F.E, McGrath, A.P, Lee, M, Guss, J.M, Mackay, J.P. | | Deposit date: | 2009-02-15 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VAC
 
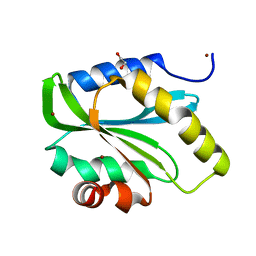 | | Structure of N-terminal Actin Depolymerizing Factor homology (ADF-H) domain of human twinfilin-2 | | Descriptor: | 1,2-ETHANEDIOL, TWINFILIN-2, ZINC ION | | Authors: | Elkins, J.M, Pike, A.C.W, King, O, Salah, E, Savitsky, P, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-08-30 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of N-Terminal Actin Depolymerizing Factor Homology (Adf-H) Domain of Human Twinfilin-2
To be Published
|
|
6QUS
 
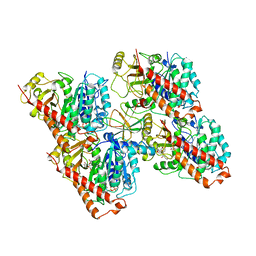 | | HsCKK (human CAMSAP1) decorated 13pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
1ZQ3
 
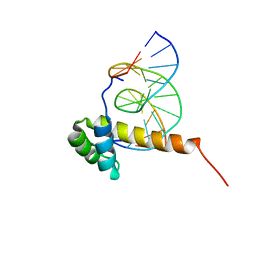 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
2V3Y
 
 | |
1ZUG
 
 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
