1WO4
 
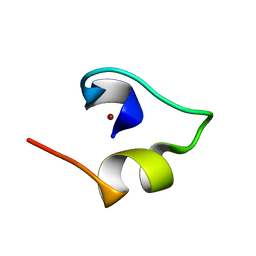 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | 分子名称: | CREB Binding Protein, ZINC ION | | 著者 | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | 登録日 | 2004-08-12 | | 公開日 | 2005-03-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WPA
 
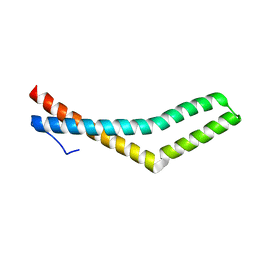 | |
8E9N
 
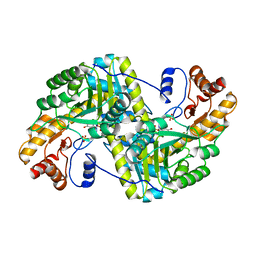 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9M
 
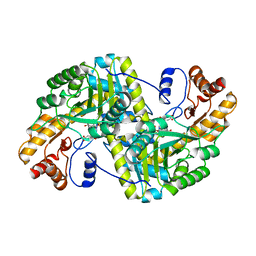 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9R
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9L
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9O
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9K
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase bound to maleate at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9T
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9Q
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9S
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9V
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
1WO5
 
 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | 分子名称: | CREB Binding Protein, ZINC ION | | 著者 | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | 登録日 | 2004-08-12 | | 公開日 | 2005-03-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1H1K
 
 | | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | | 分子名称: | RNA | | 著者 | Diprose, J.M, Grimes, J.M, Sutton, G.C, Burroughs, J.N, Meyer, A, Maan, S, Mertens, P.P.C, Stuart, D.I. | | 登録日 | 2002-07-17 | | 公開日 | 2002-09-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (10 Å) | | 主引用文献 | The Core of Bluetongue Virus Binds Double-Stranded RNA
J.Virol., 76, 2002
|
|
1X3W
 
 | | Structure of a peptide:N-glycanase-Rad23 complex | | 分子名称: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | 著者 | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | 登録日 | 2005-05-11 | | 公開日 | 2005-06-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5CIN
 
 | | Crystal Structure of non-neutralizing version of 4E10 (DeltaLoop) with epitope bound | | 分子名称: | CHLORIDE ION, FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN, ... | | 著者 | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | 登録日 | 2015-07-13 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
8E9J
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9P
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
4DD8
 
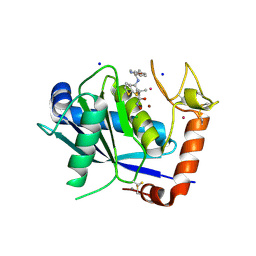 | | ADAM-8 metalloproteinase domain with bound batimastat | | 分子名称: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Hall, T, Shieh, H.S, Day, J.E, Caspers, N, Chrencik, J.E, Williams, J.M, Pegg, L.E, Pauley, A.M, Moon, A.F, Krahn, J.M, Fischer, D.H, Kiefer, J.R, Tomasselli, A.G, Zack, M.D. | | 登録日 | 2012-01-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of human ADAM-8 catalytic domain complexed with batimastat.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8E9C
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS in the ligand-free form at 100 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9D
 
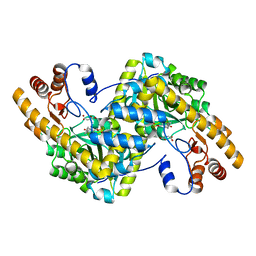 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS bound to maleic acid at 100 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9U
 
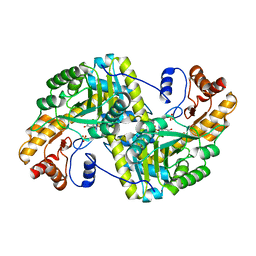 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
1WLR
 
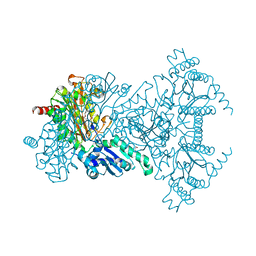 | | Apo aminopeptidase P from E. coli | | 分子名称: | CHLORIDE ION, ISOPROPYL ALCOHOL, TETRAETHYLENE GLYCOL, ... | | 著者 | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | 登録日 | 2004-06-29 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and functional implications of metal ion selection in aminopeptidase p, a metalloprotease with a dinuclear metal center
Biochemistry, 44, 2005
|
|
1WL9
 
 | | Structure of aminopeptidase P from E. coli | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, Xaa-Pro aminopeptidase | | 著者 | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | 登録日 | 2004-06-22 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional implications of metal ion selection in aminopeptidase p, a metalloprotease with a dinuclear metal center
Biochemistry, 44, 2005
|
|
8EDI
 
 | | Structure of C. elegans UNC-5 IG 1+2 Domains bound to Heparin dp4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Netrin receptor unc-5 | | 著者 | Priest, J.M, Ozkan, E. | | 登録日 | 2022-09-04 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv, 10, 2024
|
|
