6QBP
 
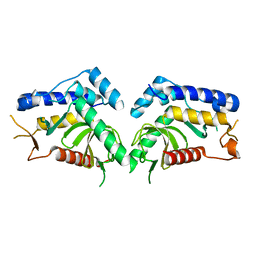 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203D | | 分子名称: | Cell wall assembly regulator SMI1 | | 著者 | Ramos, N, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | 登録日 | 2018-12-21 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203D
To Be Published
|
|
6QK4
 
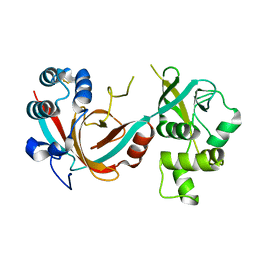 | | Lytic transglycosylase, LtgG, of Burkholderia pseudomallei. | | 分子名称: | Membrane-bound lytic murein transglycosylase A | | 著者 | Jenkins, C.H, Wallis, R, Allcock, N, Barnes, K.B, Richards, M.I, Auty, J.M, Galyov, E.E, Harding, S.V, Mukamolova, G.V. | | 登録日 | 2019-01-28 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | The lytic transglycosylase, LtgG, controls cell morphology and virulence in Burkholderia pseudomallei.
Sci Rep, 9, 2019
|
|
6QGI
 
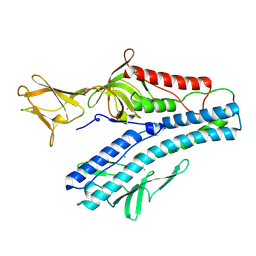 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, VP5 | | 著者 | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | 登録日 | 2019-01-11 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
6QGL
 
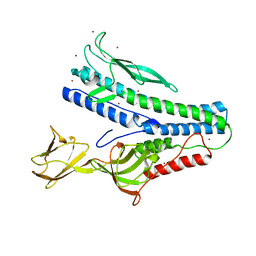 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 6 | | 分子名称: | BROMIDE ION, VP5 | | 著者 | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | 登録日 | 2019-01-11 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
4Z3U
 
 | | PRV nuclear egress complex | | 分子名称: | CHLORIDE ION, SODIUM ION, UL31, ... | | 著者 | Bigalke, J.M, Heldwein, E.E. | | 登録日 | 2015-03-31 | | 公開日 | 2015-11-11 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.706 Å) | | 主引用文献 | Structural basis of membrane budding by the nuclear egress complex of herpesviruses.
Embo J., 34, 2015
|
|
8CPK
 
 | |
6R3P
 
 | |
7R4Q
 
 | | The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.29, ... | | 著者 | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | 登録日 | 2022-02-09 | | 公開日 | 2022-06-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4I
 
 | | The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 2.15, ... | | 著者 | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | 登録日 | 2022-02-08 | | 公開日 | 2022-06-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4R
 
 | | The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.10, ... | | 著者 | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | 登録日 | 2022-02-09 | | 公開日 | 2022-06-08 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7RPC
 
 | | X-ray crystal structure of OXA-24/40 K84D in complex with ertapenem | | 分子名称: | (1S,4R,5S,6S)-3-{[(3S,5S)-5-carbamoylpyrrolidin-3-yl]sulfanyl}-6-[(1R)-1-hydroxyethyl]-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RPF
 
 | | X-ray crystal structure of OXA-24/40 in complex with doripenem | | 分子名称: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, ... | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RPG
 
 | | X-ray crystal structure of OXA-24/40 K84D in complex with cefotaxime | | 分子名称: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RPD
 
 | | X-ray crystal structure of OXA-24/40 V130D in complex with ertapenem | | 分子名称: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RPE
 
 | | X-ray crystal structure of OXA-24/40 in complex with ertapenem | | 分子名称: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RPB
 
 | | X-ray crystal structure of OXA-24/40 V130D in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RPA
 
 | | X-ray crystal structure of OXA-24/40 K84D in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
7RP9
 
 | | X-ray crystal structure of OXA-24/40 V130D in complex with imipenem | | 分子名称: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | 著者 | Powers, R.A, Mitchell, J.M, June, C.M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
4Z4B
 
 | |
7RP8
 
 | |
7R6R
 
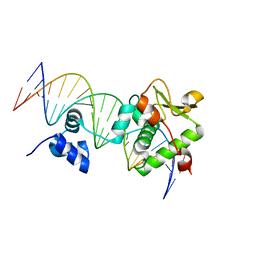 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | 分子名称: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | 著者 | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | 登録日 | 2021-06-23 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
6P59
 
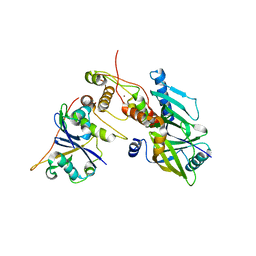 | | Crystal structure of SIVrcm Vif-CBFbeta-ELOB-ELOC complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Core-binding factor subunit beta, Elongin-B, ... | | 著者 | Binning, J.M, Chesarino, N.M, Emerman, M, Gross, J.D. | | 登録日 | 2019-05-29 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.942214 Å) | | 主引用文献 | Structural Basis for a Species-Specific Determinant of an SIV Vif Protein toward Hominid APOBEC3G Antagonism.
Cell Host Microbe, 26, 2019
|
|
1A94
 
 | | STRUCTURAL BASIS FOR SPECIFICITY OF RETROVIRAL PROTEASES | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | 著者 | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | 登録日 | 1998-04-16 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
7MGP
 
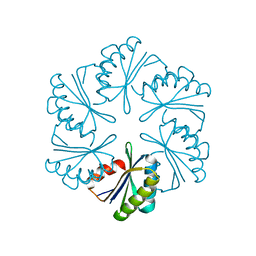 | | CmcA from Type II Cut MCP | | 分子名称: | BMC domain-containing protein | | 著者 | Ochoa, J.M, Escoto, X, Sawaya, M.R, Yeates, T.O. | | 登録日 | 2021-04-13 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MPX
 
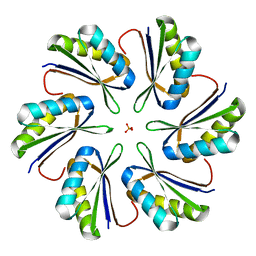 | | CmcC E36G mutant from Type II Cut MCP | | 分子名称: | BMC domain-containing protein, PHOSPHATE ION | | 著者 | Ochoa, J.M, Acosta, A.A, Sawaya, M.R, Yeates, T.O. | | 登録日 | 2021-05-05 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural characterization of hexameric shell proteins from two types of choline-utilization bacterial microcompartments
Acta Crystallogr.,Sect.F, 77, 2021
|
|
