6P5X
 
 | | Sirohydrochlorin-bound S. typhimurium siroheme synthase | | 分子名称: | 3,3',3'',3'''-[(7S,8S,12S,13S)-3,8,13,17-tetrakis(carboxymethyl)-8,13-dimethyl-7,8,12,13-tetrahydroporphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Pennington, J.M, Stroupe, M.E. | | 登録日 | 2019-05-31 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
5QD3
 
 | | Crystal structure of BACE complex with BMC010 | | 分子名称: | (10R,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4XTI
 
 | | Structure of IMP dehydrogenase of Ashbya gossypii with IMP bound to the active site | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, POTASSIUM ION | | 著者 | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | 登録日 | 2015-01-23 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
5QCY
 
 | | Crystal structure of BACE complex with BMC008 | | 分子名称: | (9R,11S)-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-16-(1,3-oxazol-2-yl)-3-[(1R)-1-phenylethyl]-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6PFW
 
 | | Protein Tyrosine Phosphatase 1B (1-301), T177A mutant, apo state | | 分子名称: | ACETATE ION, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Cui, D.S, Lipchock, J.M, Loria, J.P. | | 登録日 | 2019-06-22 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
6PJJ
 
 | | Human PRPF4B bound to benzothiophene inhibitor 224 | | 分子名称: | 1,2-ETHANEDIOL, 4-(5-{[(3-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, PHOSPHATE ION, ... | | 著者 | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-28 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | to be published
To Be Published
|
|
5U9V
 
 | | Ocellatin-LB1, solution structure in DPC micelle by NMR spectroscopy | | 分子名称: | Ocellatin-LB1 | | 著者 | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | 登録日 | 2016-12-18 | | 公開日 | 2017-03-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ocellatin-LB1
To Be Published
|
|
6AY3
 
 | | CREBBP bromodomain in complex with Cpd16 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1H-indole-3-carboxamide) | | 分子名称: | 1,2-ETHANEDIOL, 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1H-indole-3-carboxamide, CREB-binding protein, ... | | 著者 | Murray, J.M. | | 登録日 | 2017-09-07 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.391 Å) | | 主引用文献 | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
4XVX
 
 | | Crystal structure of an acyl-ACP dehydrogenase | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acyl-[acyl-carrier-protein] dehydrogenase MbtN, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | 著者 | Chai, A, Johnston, J.M, Bunker, R.D, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2015-01-28 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A covalent adduct of MbtN, an acyl-ACP dehydrogenase from Mycobacterium tuberculosis, reveals an unusual acyl-binding pocket.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6PK7
 
 | |
7KV9
 
 | | Chimeric flavivirus between Binjari virus and West Nile (Kunjin) virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Matrix protein M, envelope protein E | | 著者 | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | 登録日 | 2020-11-27 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7KVA
 
 | | Structure of West Nile virus (Kunjin) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | 著者 | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | 登録日 | 2020-11-27 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
5QCX
 
 | | Crystal structure of BACE complex with BMC007 | | 分子名称: | (9R,11S)-3-ethyl-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCP
 
 | | Crystal structure of BACE complex with BMC018 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(2-oxopropoxy)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD5
 
 | | Crystal structure of BACE complex with BMC009 | | 分子名称: | (10S,12S)-17-chloro-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-10-methyl-7-oxa-2,13,18-triazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD9
 
 | | Crystal structure of BACE complex with BMC005 | | 分子名称: | (5S,8S,10R)-8-[(1R)-2-{[1-(3-tert-butylphenyl)cyclopropyl]amino}-1-hydroxyethyl]-4,5,10-trimethyl-1-oxa-4,7-diazacyclohexadecane-3,6-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6PDI
 
 | | Human PIM1 bound to benzothiophene inhibitor 224 | | 分子名称: | 1,2-ETHANEDIOL, 4-(5-{[(3-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, GLYCEROL, ... | | 著者 | Godoi, P.H.C, Fala, A.M, Santiago, A.S, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-19 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Human PIM1
To Be Published
|
|
6P9W
 
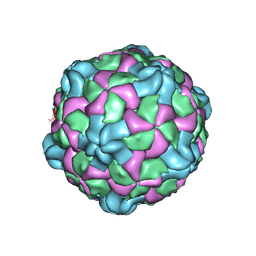 | | Poliovirus (Type 1 Mahoney), receptor catalysed 135S particle map | | 分子名称: | VP1, VP2, VP3 | | 著者 | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | 登録日 | 2019-06-10 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
7KV8
 
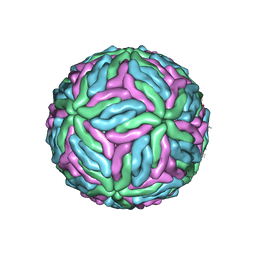 | | Chimeric flavivirus between Binjari virus and Dengue virus serotype-2 | | 分子名称: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | 著者 | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | 登録日 | 2020-11-27 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-11-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
4XUD
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd32 ([1-(biphenyl-3-yl)-5-hydroxy-4-oxo-1,4-dihydropyridin-3-yl]boronic acid) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, MAGNESIUM ION, ... | | 著者 | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | 登録日 | 2015-01-25 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4XUL
 
 | | Crystal structure of M. chilensis Mg662 protein complexed with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Lartigue, A, Priet, S, Claverie, J.M, Abergel, C. | | 登録日 | 2015-01-26 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystal structure of M. chilensis Mg662 protein complexed with GTP
To Be Published
|
|
5KC4
 
 | | Structure of TmRibU, orthorhombic crystal form | | 分子名称: | RIBOFLAVIN, Riboflavin transporter RibU, nonyl beta-D-glucopyranoside | | 著者 | Karpowich, N.K, Wang, D.N, Song, J.M. | | 登録日 | 2016-06-04 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin.
J.Mol.Biol., 428, 2016
|
|
6PM8
 
 | | Protein Tyrosine Phosphatase 1B (1-301), P180A mutant, vanadate bound state | | 分子名称: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | 著者 | Cui, D.S, Lipchock, J.M, Loria, J.P. | | 登録日 | 2019-07-01 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
6PG0
 
 | | Protein Tyrosine Phosphatase 1B (1-301), P188A mutant, vanadate bound state | | 分子名称: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | 著者 | Cui, D.S, Lipchock, J.M, Loria, J.P. | | 登録日 | 2019-06-23 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
5QCU
 
 | | Crystal structure of BACE complex with BMC022 | | 分子名称: | (2R,4S)-N-butyl-4-[(5S,8S,10R)-5,10-dimethyl-3,3,6-trioxo-3lambda~6~-thia-7-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-8-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
