1UVJ
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 7nt RNA | | 分子名称: | 5'-R(*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | 著者 | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | 登録日 | 2004-01-21 | | 公開日 | 2004-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UO0
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | 分子名称: | GENERAL CONTROL PROTEIN GCN4 | | 著者 | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | 登録日 | 2003-09-15 | | 公開日 | 2004-10-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UKH
 
 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | 分子名称: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, Mitogen-activated protein kinase 8 isoform 4 | | 著者 | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | 登録日 | 2003-08-23 | | 公開日 | 2004-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
1UO5
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | 分子名称: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, iodobenzene | | 著者 | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y. | | 登録日 | 2003-09-15 | | 公開日 | 2004-10-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UKI
 
 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | 分子名称: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, mitogen-activated protein kinase 8 isoform 4 | | 著者 | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | 登録日 | 2003-08-23 | | 公開日 | 2004-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
6SI9
 
 | | FtsZ-refold | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Cell division protein FtsZ, ... | | 著者 | Fernandez-Tornero, C, Andreu, J.M, Ruiz, F.M. | | 登録日 | 2019-08-09 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
6SOR
 
 | |
8QZP
 
 | | Structure of the non-mitochondrial citrate synthase from Ananas comosus | | 分子名称: | Citrate synthase | | 著者 | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | 登録日 | 2023-10-28 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.15 Å) | | 主引用文献 | Frequent transitions in self-assembly across the evolution of a central metabolic enzyme.
Biorxiv, 2024
|
|
8QB7
 
 | |
8QBD
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10) | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sphingolipid long chain base-responsive protein PIL1 | | 著者 | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | 登録日 | 2023-08-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | 著者 | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | 登録日 | 2023-08-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QBG
 
 | |
8QBE
 
 | |
8QB9
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30) | | 分子名称: | Sphingolipid long chain base-responsive protein PIL1 | | 著者 | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | 登録日 | 2023-08-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
5QD4
 
 | | Crystal structure of BACE complex with BMC023 | | 分子名称: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.112 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
8QB8
 
 | |
8QBB
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10) | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Sphingolipid long chain base-responsive protein PIL1 | | 著者 | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | 登録日 | 2023-08-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
6SZV
 
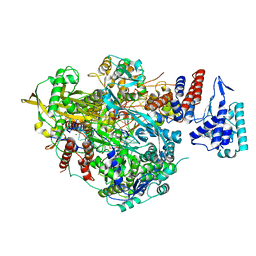 | | Bat Influenza A polymerase elongation complex with incoming UTP analogue (core + endonuclease only) | | 分子名称: | 3' vRNA, 5' vRNA, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, ... | | 著者 | Wandzik, J.M, Kouba, T, Cusack, S. | | 登録日 | 2019-10-02 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6SOP
 
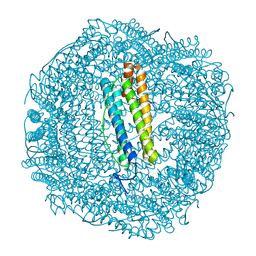 | |
6SUY
 
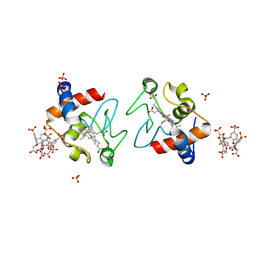 | |
6SZU
 
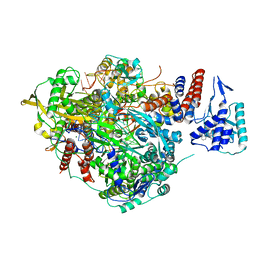 | | Bat Influenza A polymerase pre-termination complex with pyrophosphate using 44-mer vRNA template with mutated oligo(U) sequence | | 分子名称: | 5-oxidanyl-4-oxidanylidene-1-[(1-pyrrolo[2,3-b]pyridin-1-ylcyclopentyl)methyl]pyridine-3-carboxylic acid, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | 著者 | Wandzik, J.M, Kouba, T, Cusack, S. | | 登録日 | 2019-10-02 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6T0V
 
 | | Bat Influenza A polymerase elongation complex with incoming UTP analogue (complete polymerase) | | 分子名称: | 3' vRNA, 5' vRNA, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, ... | | 著者 | Wandzik, J.M, Kouba, T, Cusack, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6T0W
 
 | | Human Influenza B polymerase recycling complex | | 分子名称: | Polymerase PB2, Polymerase acidic protein, RNA-directed RNA polymerase catalytic subunit, ... | | 著者 | Wandzik, J.M, Kouba, T, Karuppasamy, M, Cusack, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
5QDB
 
 | | Crystal structure of BACE complex with BMC002 | | 分子名称: | (3S,14R,16S)-16-[(1R)-1-hydroxy-2-{[3-(1-methylethyl)benzyl]amino}ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5- dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6V3W
 
 | | Human Poly(ADP-Ribose) Polymerase 12, Catalytic fragment with four point mutations in complex with RBN-2397 | | 分子名称: | 5-{[(2S)-1-(3-oxo-3-{4-[5-(trifluoromethyl)pyrimidin-2-yl]piperazin-1-yl}propoxy)propan-2-yl]amino}-4-(trifluoromethyl)pyridazin-3(2H)-one, CHLORIDE ION, Protein mono-ADP-ribosyltransferase PARP12 | | 著者 | Swinger, K.K, Gozgit, J.M, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | 登録日 | 2019-11-26 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity.
Cancer Cell, 39, 2021
|
|
