3RCZ
 
 | | Rad60 SLD2 Ubc9 Complex | | Descriptor: | DNA repair protein rad60, SUMO-conjugating enzyme ubc9 | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
6I05
 
 | |
6HWL
 
 | | Glucosamine kinase in complex with glucosamine, ADP and inorganic phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2018-10-12 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
Mbio, 10, 2019
|
|
5NM3
 
 | | Deinococcus radiodurans BphP PAS-GAF-PHY Y263F mutant, pre-illuminated | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Takala, H, Westehoff, S, Ihalainen, J.A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome.
J. Biol. Chem., 293, 2018
|
|
6GI4
 
 | |
6GU2
 
 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUB
 
 | | CDK2/CyclinA in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GL8
 
 | | Crystal structure of Bcl-2 in complex with the novel orally active inhibitor S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Casara, P, Davidson, J, Claperon, A, Le Toumelin-Braizat, G, Vogler, M, Bruno, A, Chanrion, M, Lysiak-Auvity, G, Le Diguarher, T, Starck, J.B, Chen, I, Whitehead, N, Graham, C, Matassova, N, Dokurno, P, Pedder, C, Wang, Y, Qiu, S, Girard, A.M, Schneider, E, Grave, F, Studeny, A, Guasconi, G, Rocchetti, F, Maiga, S, Henlin, J.M, Colland, F, Kraus-Berthier, L, Le Gouill, S, Dyer, M.J.S, Hubbard, R, Wood, M, Amiot, M, Cohen, G.M, Hickman, J.A, Morris, E, Murray, J, Geneste, O. | | Deposit date: | 2018-05-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth.
Oncotarget, 9, 2018
|
|
5MF0
 
 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
6BYR
 
 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PKAc alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha chimera, MAGNESIUM ION, ... | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.661 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
6GU3
 
 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GXX
 
 | | Fab fragment of an antibody selective for alpha-1-antitrypsin in the native conformation | | Descriptor: | FAB 1D9 heavy chain, FAB 1D9 light chain, MAGNESIUM ION | | Authors: | Elliston, E.L.K, Miranda, E, Perez, J, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterisation of a monoclonal antibody conformationally-selective for native alpha-1-antitrypsin
To Be Published
|
|
6GY4
 
 | | Crystal structure of the N-terminal KH domain of human BICC1 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, CHLORIDE ION, Protein bicaudal C homolog 1, ... | | Authors: | Newman, J.A, Katis, V.L, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of the N-terminal KH domain of human BICC1
To Be Published
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
6BQ1
 
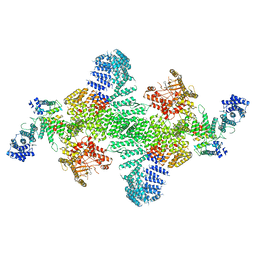 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6HMK
 
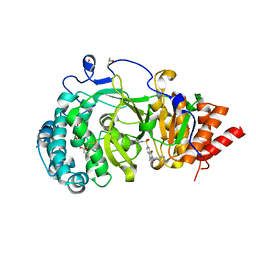 | | POLYADPRIBOSYL GLYCOHYDROLASE IN COMPLEX WITH PDD00016690 | | Descriptor: | 1-methyl-~{N}-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tucker, J.A, Barkauskaite, E. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6I0N
 
 | |
5NFG
 
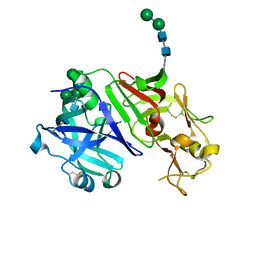 | | Structure of recombinant cardosin B from Cynara cardunculus | | Descriptor: | Procardosin-B,Procardosin-B, alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pereira, P.J.B, Figueiredo, A.C, Manso, J.A, Almeida, C.M, Simoes, I. | | Deposit date: | 2017-03-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Functional and structural characterization of synthetic cardosin B-derived rennet.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
4CGP
 
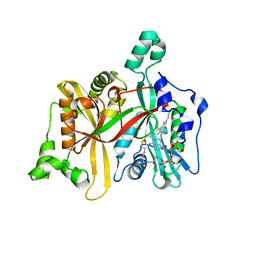 | | Leishmania major N-myristoyltransferase in complex with cofactor | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, TETRADECANOYL-COA | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
4CYO
 
 | | Leishmania major N-myristoyltransferase in complex with a hybrid inhibitor (compound 21). | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, N-{5-[(3S,4R)-1-[(3R)-3-amino-4-(4-chlorophenyl)butanoyl]-4-(hydroxymethyl)pyrrolidin-3-yl]-2-chlorophenyl}-2-(4-fluorophenyl)acetamide, ... | | Authors: | Hutton, J.A, Goncalves, V, Brannigan, J.A, Paape, D, Waugh, T, Roberts, S.M, Bell, A.S, Wilkinson, A.J, Smith, D.F, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2014-04-14 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design of Potent and Selective Leishmania N- Myristoyltransferase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4CGM
 
 | | Leishmania major N-myristoyltransferase in complex with a biphenyl- derivative inhibitor | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, N-[[3-[3-(6,7-dihydro-4H-[1,3]thiazolo[5,4-c]pyridin-5-ylmethyl)phenyl]phenyl]methyl]-2-pyridin-3-yl-ethanamine, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
4CGO
 
 | | Leishmania major N-myristoyltransferase in complex with a thienopyrimidine inhibitor | | Descriptor: | 3-[methyl-[2-[methyl-(1-methylpiperidin-4-yl)amino]thieno[3,2-d]pyrimidin-4-yl]amino]propanenitrile, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
5NBU
 
 | |
6BWZ
 
 | | SYSGYS from low-complexity domain of FUS, residues 37-42 | | Descriptor: | SYSGYS peptide from low-complexity domain of FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Tamir, G, Eisenberg, D.S. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
5NFJ
 
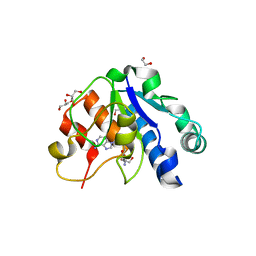 | | Crystal structure of the methyltransferase subunit of human mitochondrial Ribonuclease P (MRPP1) bound to S-adenosyl-methionine (SAM) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mitochondrial ribonuclease P protein 1, ... | | Authors: | Oerum, S, Kopec, J, Fitzpatrick, F, Newman, J.A, Chalk, R, Shrestha, L, Fairhead, M, Talon, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, C, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-14 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the methyltransferase subunit of human mitochondrial Ribonuclease P (MRPP1) bound to S-adenosyl-methionine (SAM)
To Be Published
|
|
