1N25
 
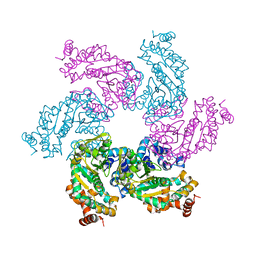 | | Crystal structure of the SV40 Large T antigen helicase domain | | Descriptor: | Large T Antigen, ZINC ION | | Authors: | Li, D, Zhao, R, Lilyestrom, W, Gai, D, Zhang, R, DeCaprio, J.A, Fanning, E, Jochimiak, A, Szakonyi, G, Chen, X.S. | | Deposit date: | 2002-10-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen
Nature, 423, 2003
|
|
1MXE
 
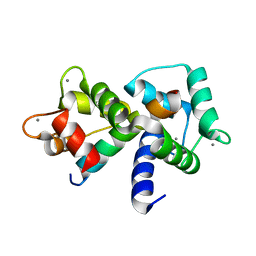 | | Structure of the Complex of Calmodulin with the Target Sequence of CaMKI | | Descriptor: | CALCIUM ION, Calmodulin, Target Sequence of rat Calmodulin-Dependent Protein Kinase I | | Authors: | Clapperton, J.A, Martin, S.R, Smerdon, S.J, Gamblin, S.J, Bayley, P.M. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Complex
of Calmodulin with the Target
Sequence of Calmodulin-Dependent
Protein Kinase I: Studies of the
Kinase Activation Mechanism
Biochemistry, 41, 2002
|
|
4EB1
 
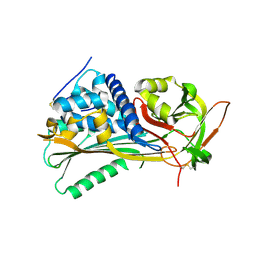 | | Hyperstable in-frame insertion variant of antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III | | Authors: | Martinez-Martinez, I, Johnson, D.J.D, Yamasaki, M, Corral, J, Huntington, J.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Type II antithrombin deficiency caused by a large in-frame insertion: structural, functional and pathological relevance.
J.Thromb.Haemost., 10, 2012
|
|
4EBP
 
 | |
1N4C
 
 | | NMR Structure of the J-Domain and Clathrin Substrate Binding Domain of Bovine Auxilin | | Descriptor: | Auxilin | | Authors: | Gruschus, J.M, Han, C.J, Greener, T, Greene, L.E, Ferretti, J.A, Eisenberg, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles.
Biochemistry, 43, 2004
|
|
4E8G
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4EC9
 
 | |
4EEN
 
 | | crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound magnesium | | Descriptor: | Beta-phosphoglucomutase-related protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4ENK
 
 | |
1J7K
 
 | | THERMOTOGA MARITIMA RUVB P216G MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1J7R
 
 | | Solution structure and backbone dynamics of the defunct EF-hand domain of Calcium Vector Protein | | Descriptor: | Calcium Vector Protein | | Authors: | Theret, I, Baladi, S, Cox, J.A, Gallay, J, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2001-05-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the defunct domain of calcium vector protein.
Biochemistry, 40, 2001
|
|
4EUN
 
 | | Crystal structure of a sugar kinase (Target EFI-502144 from Janibacter sp. HTCC2649), unliganded structure | | Descriptor: | SULFATE ION, thermoresistant glucokinase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502144 from Janibacter sp. HTCC2649), unliganded structure
To be Published
|
|
4EBU
 
 | | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-24 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I
TO BE PUBLISHED
|
|
4E4F
 
 | | Crystal structure of enolase PC1_0802 (TARGET EFI-502240) from Pectobacterium carotovorum subsp. carotovorum PC1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ENOLASE PC1_0802 from Pectobacterium carotovorum
To be Published
|
|
4ECI
 
 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with acetate | | Descriptor: | ACETATE ION, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
4FRE
 
 | | Crystal Structure of BBBB+UDP+Gal at pH 6.5 with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
4FRM
 
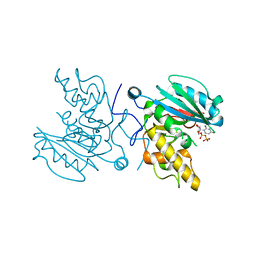 | | Crystal Structure of BBBB+UDP+Gal at pH 7.0 with MPD as the cryoprotectant | | Descriptor: | Histo-blood group ABO system transferase, URIDINE-5'-DIPHOSPHATE, beta-D-galactopyranose | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
4FKL
 
 | |
1IHV
 
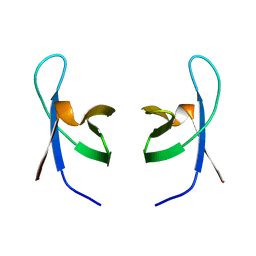 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
4FSN
 
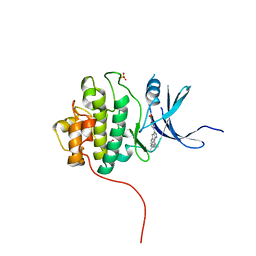 | | Crystal Structure of the CHK1 | | Descriptor: | 4-(6-{[(4-METHYLCYCLOHEXYL)AMINO]METHYL}-1,4-DIHYDROINDENO[1,2-C]PYRAZOL-3-YL)BENZOIC ACID, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FUB
 
 | | Crystal Structure of the Urokinase | | Descriptor: | 6-[(2S,3S)-3-phenyloxiran-2-yl]naphthalene-2-carboximidamide, ACETATE ION, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Urokinase
to be published
|
|
4G4M
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3(6-13) | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, alpha4F3(6-13) | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
4FV2
 
 | | Crystal Structure of the ERK2 complexed with EK5 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-chlorophenyl)-1H-pyrazol-5-yl]-N-(2,3-dihydro-1-benzofuran-5-ylmethyl)-1H-pyrrole-2-carboxamide, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the ERK2 complexed with EK5
TO BE PUBLISHED
|
|
4FX6
 
 | | Crystal structure of the mutant V182A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-02 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4GAY
 
 | | Structure of the broadly neutralizing antibody AP33 | | Descriptor: | NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, NEUTRALIZING ANTIBODY AP33 LIGHT CHAIN, TRIETHYLENE GLYCOL | | Authors: | Potter, J.A, Owsianka, A, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-10 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
