1T3S
 
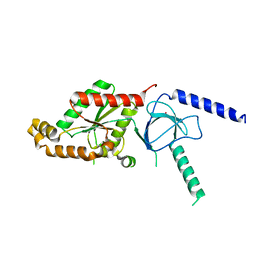 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, MERCURY (II) ION | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain.
Neuron, 42, 2004
|
|
1GUO
 
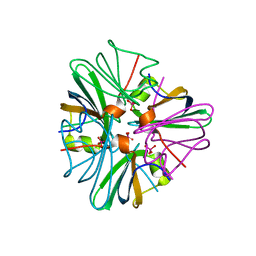 | |
1GUS
 
 | |
8DO6
 
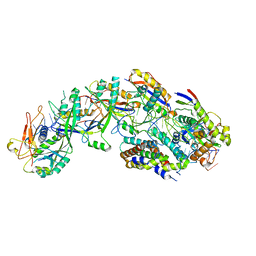 | | The structure of S. epidermidis Cas10-Csm bound to target RNA | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Paraan, M, Stagg, S.M, Dunkle, J.A. | | Deposit date: | 2022-07-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems.
Plos One, 18, 2023
|
|
1GUN
 
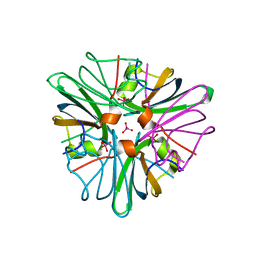 | |
3N38
 
 | | Ribonucleotide Reductase NrdF from Escherichia coli Soaked with Ferrous Ions | | Descriptor: | FE (II) ION, Ribonucleoside-diphosphate reductase 2 subunit beta | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
7MK0
 
 | |
1GUT
 
 | |
8EUL
 
 | | cytochrome P450terp (cyp108A1) mutant F188A bound to alpha-terpineol | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450-terp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gable, J.A, Follmer, A.H, Poulos, T.L. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Cooperative Substrate Binding Controls Catalysis in Bacterial Cytochrome P450terp (CYP108A1).
J.Am.Chem.Soc., 2023
|
|
8ESW
 
 | | Structure of mitochondrial complex I from Drosophila melanogaster, Flexible-class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Padavannil, A, Letts, J.A. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Resting mitochondrial complex I from Drosophila melanogaster adopts a helix-locked state.
Elife, 12, 2023
|
|
8ESZ
 
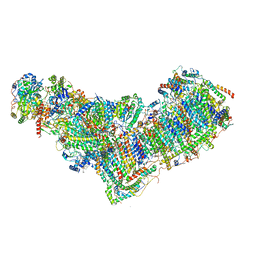 | | Structure of mitochondrial complex I from Drosophila melanogaster, Helix-locked state | | Descriptor: | (2R)-3-{[(S)-hydroxy(3-methylbutoxy)phosphoryl]oxy}-2-(octanoyloxy)propyl decanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Padavannil, A, Letts, J.A. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Resting mitochondrial complex I from Drosophila melanogaster adopts a helix-locked state.
Elife, 12, 2023
|
|
1GUG
 
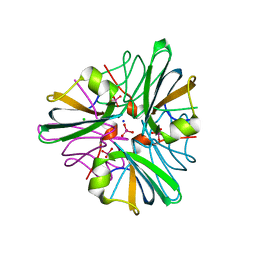 | | MopII from Clostridium pasteurianum complexed with tungstate | | Descriptor: | CHLORIDE ION, MOLYBDATE BINDING PROTEIN II, SODIUM ION, ... | | Authors: | Schuettelkopf, A.W, Harrison, J.A, Hunter, W.N. | | Deposit date: | 2002-01-25 | | Release date: | 2002-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Passive Acquisition of Ligand by the Mopii Molbindin from Clostridium Pasteurianum: Structures of Apo and Oxyanion-Bound Forms
J.Biol.Chem., 277, 2002
|
|
1H2G
 
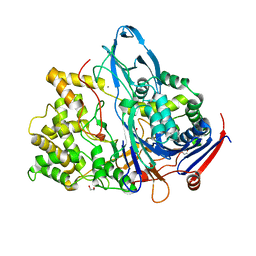 | | Altered substrate specificity mutant of penicillin acylase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Morillas, M, Brannigan, J.A, Ladurner, A.G, Forney, L.J, Virden, R. | | Deposit date: | 2002-08-08 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of Penicillin Acylase Residue B71 Extend Substrate Specificity by Decreasing Steric Constraints for Substrate Binding
Biochem.J., 371, 2003
|
|
3NOD
 
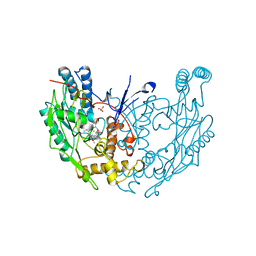 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND PRODUCT ANALOGUE L-THIOCITRULLINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, L-THIOCITRULLINE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
1T3L
 
 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core in Complex with Alpha1 Interaction Domain | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, Voltage-dependent L-type calcium channel alpha-1S subunit | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Voltage-Dependent Calcium Channel Beta Subunit Functional Core and Its Complex with the Alpha1 Interaction Domain
NEURON, 42, 2004
|
|
1VND
 
 | | VND/NK-2 PROTEIN (HOMEODOMAIN), NMR | | Descriptor: | VND/NK-2 PROTEIN | | Authors: | Tsao, D.H.H, Gruschus, J.M, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1996-05-22 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the NK-2 homeodomain from Drosophila.
J.Mol.Biol., 251, 1995
|
|
8C0S
 
 | |
8C0P
 
 | |
1FSE
 
 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS REGULATORY PROTEIN GERE | | Descriptor: | GERE, GLYCEROL, SULFATE ION | | Authors: | Ducros, V.M.-A, Lewis, R.J, Verma, C.S, Dodson, E.J, Leonard, G, Turkenburg, J.P, Murshudov, G.N, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2000-09-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis.
J.Mol.Biol., 306, 2001
|
|
8CF3
 
 | |
1FLJ
 
 | | CRYSTAL STRUCTURE OF S-GLUTATHIOLATED CARBONIC ANHYDRASE III | | Descriptor: | CARBONIC ANHYDRASE III, GLUTATHIONE, ZINC ION | | Authors: | Mallis, R.J, Poland, B.W, Chatterjee, T.K, Fisher, R.A, Darmawan, S, Honzatko, R.B, Thomas, J.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of S-glutathiolated carbonic anhydrase III.
FEBS Lett., 482, 2000
|
|
8ELU
 
 | | HRAS R97G Crystal form 1 | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Rodrigues, J.A, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
8EML
 
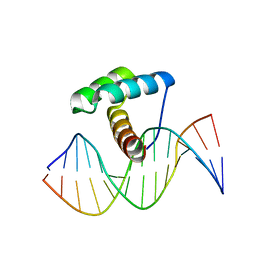 | | Crystal Structure of Gsx2 Homeodomain in Complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*GP*CP*TP*AP*AP*TP*TP*AP*AP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*TP*TP*AP*AP*TP*TP*AP*GP*CP*TP*C)-3'), GS homeobox 2, ... | | Authors: | Webb, J.A, Kovall, R.A. | | Deposit date: | 2022-09-28 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Gsx2 Homeodomain in Complex with DNA
To Be Published
|
|
1GNX
 
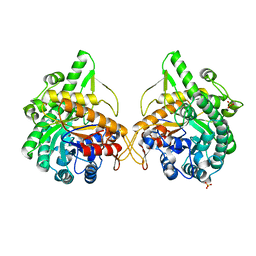 | | b-glucosidase from Streptomyces sp | | Descriptor: | BETA-GLUCOSIDASE, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Guasch, A, Perez-Pons, J.A, Vallmitjana, M, Querol, E, Coll, M. | | Deposit date: | 2001-10-10 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Beta-Glucosidase from Streptomyces
To be Published
|
|
8EUH
 
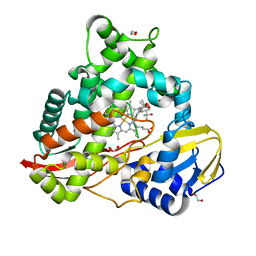 | | cytochrome P450terp (cyp108A1) bound to alpha-terpineol | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450-terp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gable, J.A, Follmer, A.H, Poulos, T.L. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative Substrate Binding Controls Catalysis in Bacterial Cytochrome P450terp (CYP108A1).
J.Am.Chem.Soc., 2023
|
|
