3G0D
 
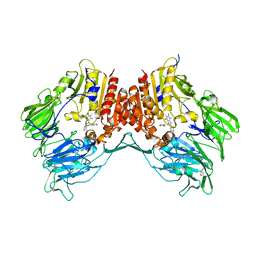 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 2 | | Descriptor: | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
3G1X
 
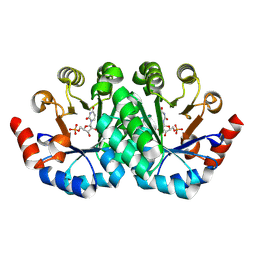 | | Crystal structure of the mutant D70G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4Y63
 
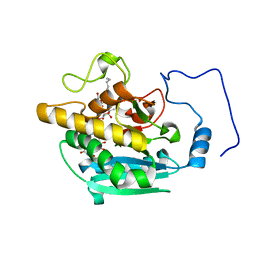 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-deoxy-5'-{[(2S)-2-(triaza-1,2-dien-2-ium-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
2W1I
 
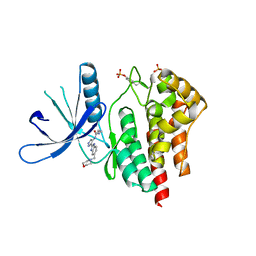 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 4-[(2-{4-[(CYCLOPROPYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-1H-BENZIMIDAZOL-6-YL)METHYL]MORPHOLIN-4-IUM, JAK2 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
2WCH
 
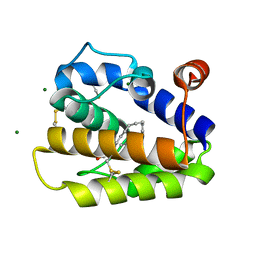 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with bombykal | | Descriptor: | (10E,12Z)-hexadeca-10,12-dienal, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
4YW5
 
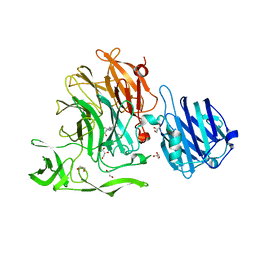 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
2X36
 
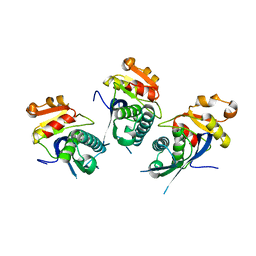 | | Structure of the proteolytic domain of the Human Mitochondrial Lon protease | | Descriptor: | LON PROTEASE HOMOLOG, MITOCHONDRIAL | | Authors: | Garcia, J, Ondrovicova, G, Blagova, E, Levdikov, V.M, Bauer, J.A, Kutejova, E, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2010-01-21 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Catalytic Domain of the Human Mitochondrial Lon Protease: Proposed Relation of Oligomer Formation and Activity.
Protein Sci., 19, 2010
|
|
2X8E
 
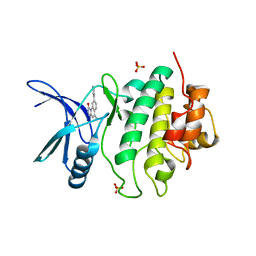 | | Discovery of a Novel Class of triazolones as Checkpoint Kinase Inhibitors - Hit to Lead Exploration | | Descriptor: | 5-METHYL-8-PYRIDIN-4-YL[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1(2H)-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Novel Class of Triazolones as Checkpoint Kinase Inhibitors-Hit to Lead Exploration.
Bioorg.Med.Chem., 20, 2010
|
|
2X8M
 
 | |
7QRL
 
 | |
7QVD
 
 | |
3FYY
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with Mg | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
2X8I
 
 | | Discovery of a Novel Class of triazolones as Checkpoint Kinase Inhibitors - Hit to Lead Exploration | | Descriptor: | 7-(3-hydroxyphenyl)-5-methyl[1,2,4]triazolo[4,3-a]quinolin-1(2H)-one, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Otterbein, L, Vallentine, A, White, A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of a Novel Class of Triazolones as Checkpoint Kinase Inhibitors-Hit to Lead Exploration.
Bioorg.Med.Chem., 20, 2010
|
|
3G1D
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4Y62
 
 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-deoxy-5'-{[(2R)-3-hydroxy-2-(4-phenyl-1H-1,2,3-triazol-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
7RG1
 
 | | Importin alpha2 in complex with MERS ORF4B H26A mutant | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Importin alpha2 in complex with MERS ORF4B NLS H26A mutant
To Be Published
|
|
7RFY
 
 | | Importin alpha3 in complex with MERS ORF4B | | Descriptor: | Importin subunit alpha-3, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
2WKA
 
 | | Structure of Plp_Thr_decanoyl-CoA aldimine form of Vibrio cholerae CqsA | | Descriptor: | CAI-1 AUTOINDUCER SYNTHASE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Jahan, N, Potter, J.A, Sheikh, M.A, Botting, C.H, Shirran, S.L, Westwood, N.J, Taylor, G.L. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Insights Into the Biosynthesis of the Vibrio Cholerae Major Autoinducer Cai-1 from the Crystal Structure of the Plp-Dependent Enzyme Cqsa.
J.Mol.Biol., 392, 2009
|
|
4YC3
 
 | | CDK1/CyclinB1/CKS2 Apo | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-19 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
7RG0
 
 | | Importin alpha2 in complex with MERS ORF4B R24A mutant | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RFX
 
 | | Importin alpha2 in complex with MERS ORF4B | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RFZ
 
 | | Importin alpha 2 in complex with MERS ORF4B NLS peptide | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
4YEY
 
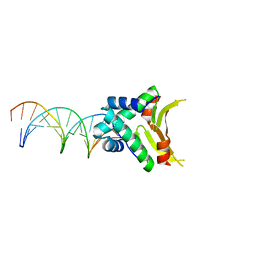 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
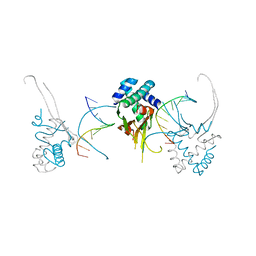 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YC8
 
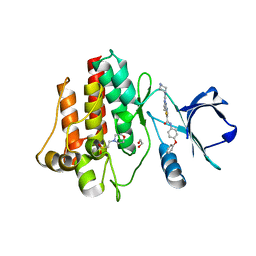 | | C-Helix-Out Binding of Dasatinib Analog to c-Abl Kinase | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(4-phenoxyphenyl)-1,3-thiazole-5-carboxamide, ... | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.J, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-19 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
