5JY4
 
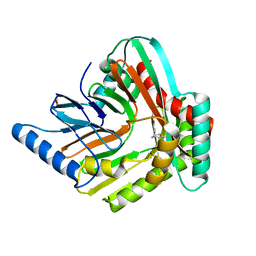 | | A high magnesium structure of the isochorismate synthase, EntC | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, MAGNESIUM ION | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
6S3Y
 
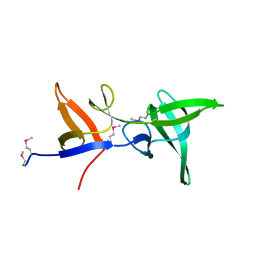 | |
6ROH
 
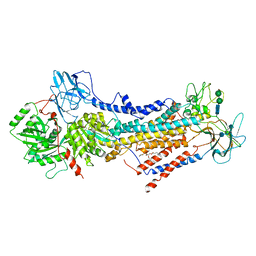 | | Cryo-EM structure of the autoinhibited Drs2p-Cdc50p | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
5LNK
 
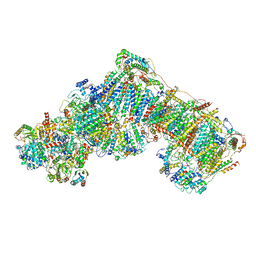 | | Entire ovine respiratory complex I | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 4'-PHOSPHOPANTETHEINE, ... | | Authors: | Fiedorczuk, K, Letts, J.A, Kaszuba, K, Sazanov, L.A. | | Deposit date: | 2016-08-04 | | Release date: | 2016-11-23 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the entire mammalian mitochondrial complex I.
Nature, 538, 2016
|
|
3MP2
 
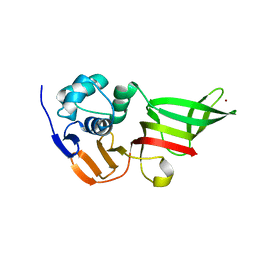 | |
5LMX
 
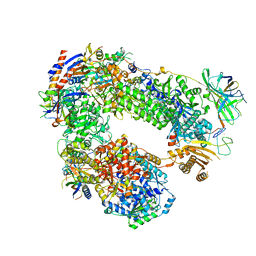 | | Monomeric RNA polymerase I at 4.9 A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Torreira, E, Louro, J.A, Gil-Carton, D, Gallego, O, Calvo, O, Fernandez-Tornero, C. | | Deposit date: | 2016-08-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription.
Elife, 6, 2017
|
|
6S33
 
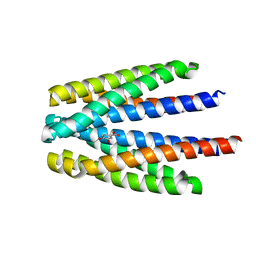 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with Protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
3M1Z
 
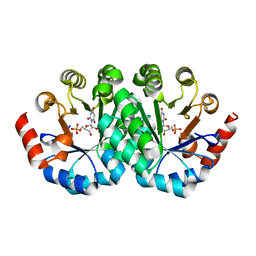 | | Crystal structure of the mutant V182A.V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3M44
 
 | | Crystal structure of the mutant V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3M5X
 
 | | Crystal structure of the mutant V182A,I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
6RIO
 
 | | Imidazole Polyamide-DNA complex NMR structure (5'-CGATGTACATCG-3') | | Descriptor: | 3-[3-[[4-[[4-[[4-[[4-[[(2~{R})-2-azaniumyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoyl]amino]-1-methyl-imidazol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]propanoylamino]propyl-dimethyl-azanium, DNA (5'-(*(DC5)P*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*(DG3))-3') | | Authors: | Padroni, G, Withers, J.M, Taladriz-Sender, A, Reichenbach, L.F, Parkinson, J.A, Burley, G.A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequence-Selective Minor Groove Recognition of a DNA Duplex Containing Synthetic Genetic Components.
J.Am.Chem.Soc., 141, 2019
|
|
5JXZ
 
 | | A low magnesium structure of the isochorismate synthase, EntC | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, ... | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
3KFE
 
 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
5JCG
 
 | |
6U9N
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 14 (TC-5139) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(4-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
3KRM
 
 | | Imp1 kh34 | | Descriptor: | GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 1 | | Authors: | Chao, J.A, Singer, R.H, Almo, S.C, Patskovsky, Y. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ZBP1 recognition of beta-actin zipcode induces RNA looping.
Genes Dev., 24, 2010
|
|
5JGY
 
 | | Crystal structure of maize AKR4C13 in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]oxy}butanoic acid (non-preferred name), Aldose reductase, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of maize AKR4C13 in P21 space group
To Be Published
|
|
6S38
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
5LD9
 
 | | Structure of deubiquitinating enzyme homolog, Pyrococcus furiosus JAMM1. | | Descriptor: | CHLORIDE ION, JAMM1, ZINC ION | | Authors: | Maupin-Furlow, J.A, Franzetti, B, Cao, S, Girard, E, Gabel, F, Engilberge, S. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-17 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
6RRC
 
 | | Crystal structure of the N-terminal region of human cohesin subunit STAG1 in complex with RAD21 peptide | | Descriptor: | Cohesin subunit SA-1, Double-strand-break repair protein rad21 homolog, SULFATE ION | | Authors: | Newman, J.A, Katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6THJ
 
 | | CBDP35 Native structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PlyP35 | | Authors: | Hermoso, J.A, Bartual, S.G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CBDP35 Native structure
To Be Published
|
|
6TT5
 
 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6TPN
 
 | | Crystal structure of the Orexin-2 receptor in complex with HTL6641 at 2.61 A resolution | | Descriptor: | 2-(5,6-dimethoxypyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, NITRATE ION, OLEIC ACID, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5JH1
 
 | | Crystal structure of the apo form of AKR4C7 from maize | | Descriptor: | Aldose reductase, AKR4C7 | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
6TOD
 
 | | Crystal structure of the Orexin-1 receptor in complex with EMPA | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
