2XPC
 
 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | Descriptor: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
2XYQ
 
 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
1JZO
 
 | | DsbC C101S | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN DSBC | | Authors: | Haebel, P.W, Goldstone, D, Katzen, F, Beckwith, J, Metcalf, P. | | Deposit date: | 2001-09-17 | | Release date: | 2003-03-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Disulfide Bond Isomerase DsbC is Activated by an
Immunoglobulin-fold Thiol Oxidoreductase: Crystal Structure of the
DsbC-DsbDalpha complex.
Embo J., 21, 2002
|
|
2Y0B
 
 | | Caspase-3 in Complex with an Inhibitory DARPin-3.4_S76R | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CASPASE-3, ... | | Authors: | Barandun, J, Schroeder, T, Mittl, P, Grutter, M.G. | | Deposit date: | 2010-12-01 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specific Inhibition of Caspase-3 by a Competitive Darpin: Molecular Mimicry between Native and Designed Inhibitors.
Structure, 21, 2013
|
|
1I78
 
 | | CRYSTAL STRUCTURE OF OUTER MEMBRANE PROTEASE OMPT FROM ESCHERICHIA COLI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PROTEASE VII, octyl beta-D-glucopyranoside | | Authors: | Vandeputte-Rutten, L, Kramer, R.A, Kroon, J, Dekker, N, Egmond, M.R, Gros, P. | | Deposit date: | 2001-03-08 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site.
EMBO J., 20, 2001
|
|
2XVD
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, {4-METHYL-3-[(1-METHYLETHYL)(2-{[3-(METHYLSULFONYL)-5-MORPHOLIN-4-YLPHENYL]AMINO}PYRIMIDIN-4-YL)AMINO]PHENYL}METHANOL | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L. | | Deposit date: | 2010-10-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 4: Discovery and Optimization of a Benzylic Alcohol Series.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6S71
 
 | | Crystal structure of CARM1 in complex with inhibitor WH5C | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(5-carbamimidamidopentyl)amino]-2-azanyl-butanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
2XWA
 
 | | Crystal Structure of Complement Factor D Mutant R202A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
6S7B
 
 | | Crystal structure of CARM1 in complex with inhibitor UM249 | | Descriptor: | 1-[4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]butyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
2ZG8
 
 | | Crystal Structure of Pd(allyl)/apo-H49AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Niemeyer, J, Abe, M, Ueno, T, Hikage, T, Erker, G, Watanabe, Y. | | Deposit date: | 2008-01-18 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Control of the coordination structure of organometallic palladium complexes in an apo-ferritin cage.
J.Am.Chem.Soc., 130, 2008
|
|
2XZZ
 
 | | Crystal structure of the human transglutaminase 1 beta-barrel domain | | Descriptor: | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE K | | Authors: | Vollmar, M, Krysztofinska, E, Krojer, T, Yue, W.W, Cooper, C, Kavanagh, K, Allerston, C, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Human Transglutaminase 1 Beta-Barrel Domain
To be Published
|
|
6RUK
 
 | |
6RUW
 
 | |
2Y0K
 
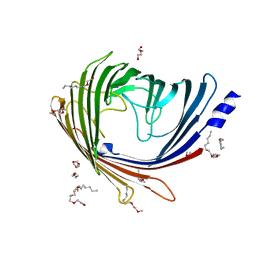 | |
6S1S
 
 | | Crystal structure of AmpC from Pseudomonas aeruginosa in complex with [3-(2-carboxyvinyl)phenyl]boronic acid] inhibitor | | Descriptor: | (~{E})-3-[3-(dihydroxyboranyl)phenyl]prop-2-enoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Kekez, I, Vicario, M, Bellio, P, Tosoni, E, Celenza, G, Blazquez, J, Tondi, D, Cendron, L. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phenylboronic Acids Probing Molecular Recognition against Class A and Class C beta-lactamases.
Antibiotics, 8, 2019
|
|
2Y2X
 
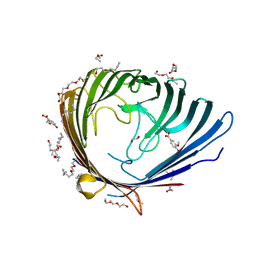 | |
6RVD
 
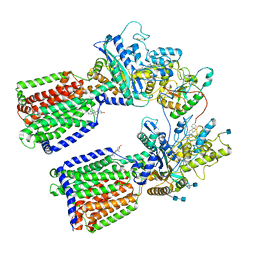 | | Revised cryo-EM structure of the human 2:1 Ptch1-Shh complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | El Omari, K, Rudolf, A.F, Kowatsch, C, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
2Y32
 
 | | Crystal structure of bradavidin | | Descriptor: | BLR5658 PROTEIN | | Authors: | Leppiniemi, J, Gronroos, T, Johnson, M.S, Kulomaa, M.S, Hytonen, V.P, Airenne, T.T. | | Deposit date: | 2010-12-17 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Bradavidin - C-Terminal Residues Act as Intrinsic Ligands.
Plos One, 7, 2012
|
|
6RW4
 
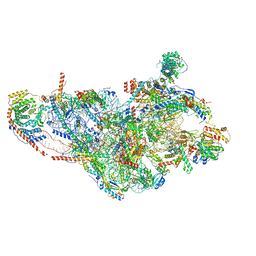 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
6S2G
 
 | | Structure Of D80A-Fructofuranosidase From Xanthophyllomyces Dendrorhous Complexed With Fructose And Epigallocatechin Gallate (Egcg) | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-06-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Deciphering the molecular specificity of phenolic compounds as inhibitors or glycosyl acceptors of beta-fructofuranosidase from Xanthophyllomyces dendrorhous.
Sci Rep, 9, 2019
|
|
2Y5K
 
 | | Orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | 1-[5-(2-METHOXYETHYL)-4-METHYL-THIOPHEN-2-YL]SULFONYL-3-[4-METHOXY-6-(METHYLCARBAMOYLAMINO)PYRIDIN-2-YL]UREA, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Hebeisen, P, Haap, W, Kuhn, B, Mohr, P, Wessel, H.P, Zutter, U, Kirchner, S, Benz, J, Joseph, C, Alvarez-Sanchez, R, Gubler, M, Schott, B, Benardeau, A, Tozzo, E, Kitas, E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1JQ4
 
 | | [2Fe-2S] Domain of Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Mueller, J, Lugovskoy, A.A, Wagner, G, Lippard, S.J. | | Deposit date: | 2001-08-03 | | Release date: | 2002-01-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the [2Fe-2S] ferredoxin domain from soluble methane monooxygenase reductase and interaction with its hydroxylase.
Biochemistry, 41, 2002
|
|
3C91
 
 | | Thermoplasma acidophilum 20S proteasome with an open gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
2XVW
 
 | |
6S7A
 
 | | Crystal structure of CARM1 in complex with inhibitor AA175 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-azanylpropyl-[3-(pyridin-2-ylamino)propyl]amino]methyl]oxolane-3,4-diol, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Al-Noori, A, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
