6TNH
 
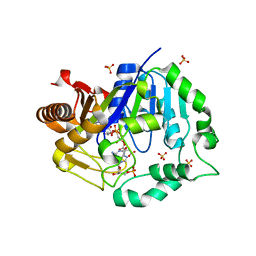 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with AMPPcP, dGMP, Asp, Magnesium at 2.21 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ASPARTIC ACID, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
1OPL
 
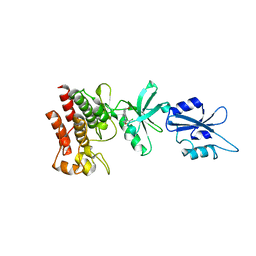 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, MYRISTIC ACID, proto-oncogene tyrosine-protein kinase | | Authors: | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
6T1Q
 
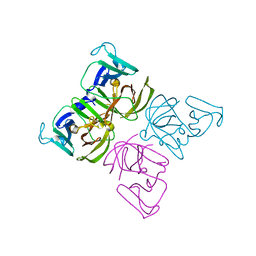 | | 3C-like protease from Southampton norovirus. | | Descriptor: | Genome polyprotein | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
2FS2
 
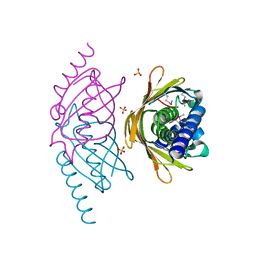 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
2FSR
 
 | | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, acetyltransferase | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Gu, J, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58
To be Published
|
|
6TQD
 
 | | D00-D0 domain of Intimin | | Descriptor: | BROMIDE ION, CHLORIDE ION, Intimin, ... | | Authors: | Weikum, J, Leo, J.C, Morth, J.P. | | Deposit date: | 2019-12-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep, 10, 2020
|
|
6T4C
 
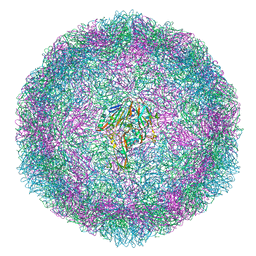 | | Bovine enterovirus F3 in complex with glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
3MXZ
 
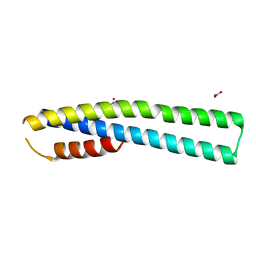 | | Crystal Structure of tubulin folding cofactor A from Arabidopsis thaliana | | Descriptor: | NITRATE ION, Tubulin-specific chaperone A | | Authors: | Lu, L, Nan, J, Mi, W, Su, X.D, Li, Y. | | Deposit date: | 2010-05-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Crystal structure of tubulin folding cofactor A from Arabidopsis thaliana and its beta-tubulin binding characterization
Febs Lett., 584, 2010
|
|
2FQ3
 
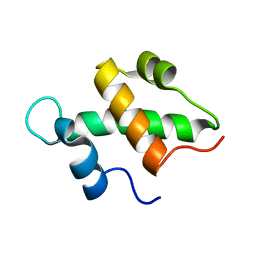 | | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | | Descriptor: | Transcription regulatory protein SWI3 | | Authors: | Da, G, Lenkart, J, Zhao, K, Shiekhattar, R, Cairns, B.R, Marmorstein, R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6TAY
 
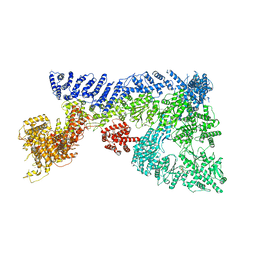 | | Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related Human variant R4810K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
6TKJ
 
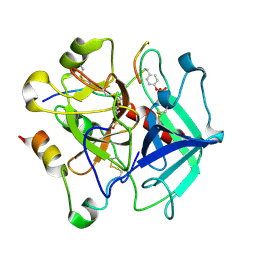 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - tetragonal form at 7keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
6TBO
 
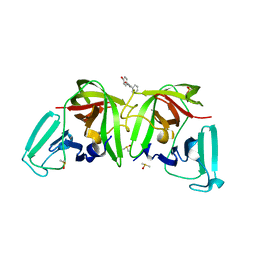 | |
6TBP
 
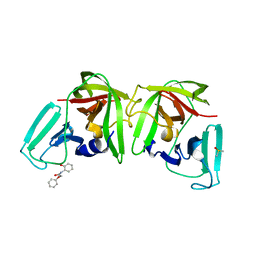 | |
2FKW
 
 | | Structure of LH2 from Rps. acidophila crystallized in lipidic mesophases | | Descriptor: | BACTERIOCHLOROPHYLL A, LAURYL DIMETHYLAMINE-N-OXIDE, Light-harvesting protein B-800/850, ... | | Authors: | Papiz, M.Z, Cherezov, V, Clogston, J, Caffrey, M. | | Deposit date: | 2006-01-05 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Room to Move: Crystallizing Membrane Proteins in Swollen Lipidic Mesophases
J.Mol.Biol., 357, 2006
|
|
1DCU
 
 | | REDOX SIGNALING IN THE CHLOROPLAST: STRUCTURE OF OXIDIZED PEA FRUCTOSE-1,6-BISPHOSPHATE PHOSPHATASE | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Chiadmi, M, Navaza, A, Miginiac-Maslow, M, Jacquot, J.P, Cherfils, J. | | Deposit date: | 1999-11-05 | | Release date: | 1999-12-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
EMBO J., 18, 1999
|
|
2FMH
 
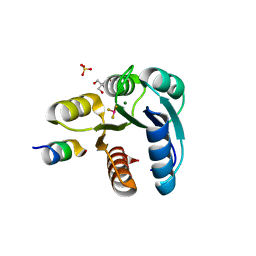 | | Crystal structure of Mg2+ and BeF3- bound CheY in complex with CheZ 200-214 solved from a F432 crystal grown in Tris (pH 8.4) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BERYLLIUM TRIFLUORIDE ION, C-terminal 15-mer from Chemotaxis protein cheZ, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation.
J.Mol.Biol., 359, 2006
|
|
2F2B
 
 | | Crystal structure of integral membrane protein Aquaporin AqpM at 1.68A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6T68
 
 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T71
 
 | |
2F3V
 
 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F4T
 
 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3', 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
6TAW
 
 | |
6T2T
 
 | | Crystal structure of Drosophila melanogaster glutathione S-transferase epsilon 14 in complex with glutathione and 2-methyl-2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Skerlova, J, Lindstrom, H, Sjodin, B, Gonis, E, Neiers, F, Mannervik, B, Stenmark, P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and steroid isomerase activity of Drosophila glutathione transferase E14 essential for ecdysteroid biosynthesis.
Febs Lett., 594, 2020
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
2FAZ
 
 | | Ubiquitin-Like Domain of Human Nuclear Zinc Finger Protein NP95 | | Descriptor: | Ubiquitin-like containing PHD and RING finger domains protein 1 | | Authors: | Walker, J.R, Wybenga-Groot, L, Doherty, R.S, Finerty Jr, P.J, Newman, E, Mackenzie, F.M, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-08 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin-Like Domain of Human Nuclear Zinc Finger Protein NP95
To be Published
|
|
