8OKT
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor VK42 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{R},9~{S},13~{S},14~{S})-13-methyl-17-oxidanylidene-7,8,9,11,12,14,15,16-octahydro-6~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8EZH
 
 | | A tethered niacin-derived pincer complex with a nickel-carbon bond in lactate racemase R98A/R100A variant modeled with sulfite-NPN adduct | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Gatreddi, S, Hausinger, R.P, Hu, J. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Irreversible inactivation of lactate racemase by sodium borohydride reveals reactivity of the nickel-pincer nucleotide cofactor.
Acs Catalysis, 13, 2023
|
|
8OLM
 
 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl AKI1 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},5~{S},8~{S},13~{R},14~{R})-5,14-dimethyl-16-oxidanylidene-1,2,3,4,6,7,8,11,12,13,15,17-dodecahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OMB
 
 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl VK42 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{R},9~{S},13~{S},14~{S})-13-methyl-17-oxidanylidene-7,8,9,11,12,14,15,16-octahydro-6~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8F0W
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
8OMN
 
 | | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl VK4 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
1FFK
 
 | | CRYSTAL STRUCTURE OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Ban, N, Nissen, P, Hansen, J, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.
Science, 289, 2000
|
|
8OZ8
 
 | | Crystal Structure of an Hydroxynitrile lyase variant (H96A) from Granulicella tundricola | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, BROMIDE ION, ... | | Authors: | Bento, I, Coloma, J, Hagedoorn, P.-L, Hanefeld, U. | | Deposit date: | 2023-05-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Can a Hydroxynitrile Lyase Catalyze an Oxidative Cleavage?
Acs Catalysis, 13, 2023
|
|
5IQO
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitutions Q134E and S138E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Giese, C, Eras, J, Kern, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8OKE
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_1 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},8~{R},9~{S},10~{R},13~{S},14~{S})-10,13-dimethyl-17-oxidanylidene-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
5KP2
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae cocrystallized with octanoyl-CoA: hydrolzed ligand | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Hou, J, Zheng, H, Grabowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: hydrolzed ligand
To Be Published
|
|
5KPO
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
8OKG
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_13 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},8~{S},9~{S},10~{R},13~{S},14~{S},17~{S})-17-methanoyl-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
5KR3
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KRC
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with Zearalenone | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5KRP
 
 | | Frutapin, a lectin from Artocarpus incisa: Cloning, Expressing and Structural Analysis. | | Descriptor: | Frutapin, GLYCEROL | | Authors: | de Sousa, F.D, Coker, A.R, Guan, Y, Guo, J, de Oliveira Monteiro-Moreira, A.C, de Azevedo Moreira, R. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Frutapin, a lectin fromArtocarpus incisa(breadfruit): cloning, expression and molecular insights.
Biosci. Rep., 37, 2017
|
|
8OUM
 
 | | Arf GTPase from the asgard Gerdarchaea : GerdArfR1 bound to GTP | | Descriptor: | GTP-binding protein, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Menetrey, J, Jackson, C, Dacks, J.B, Elias, M, Vargova, R. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Arf Family GTPases are present in Asgard archaea
To Be Published
|
|
7V9Z
 
 | |
8UO6
 
 | |
1M74
 
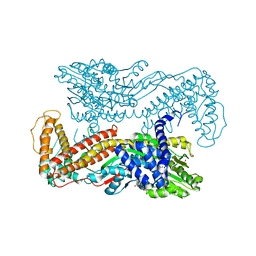 | | Crystal structure of Mg-ADP-bound SecA from Bacillus subtilis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase secA, ... | | Authors: | Hunt, J.F, Weinkauf, S, Henry, L, Fak, J.J, McNicholas, P, Oliver, D.B, Deisenhofer, J. | | Deposit date: | 2002-07-16 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA
Science, 297, 2002
|
|
8CBJ
 
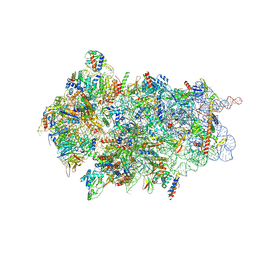 | | Cryo-EM structure of Otu2-bound cytoplasmic pre-40S ribosome biogenesis complex | | Descriptor: | 20S pre-ribosomal RNA, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8C4X
 
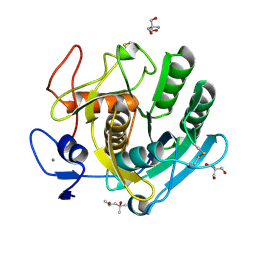 | | PAM Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Nomme, J, Gavalda, S, Cioci, G, Guicherd, M, Marty, A, Duquesne, S, Ben Khaled, M. | | Deposit date: | 2023-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | An engineered enzyme embedded into PLA to make self-biodegradable plastic.
Nature, 631, 2024
|
|
8C4Z
 
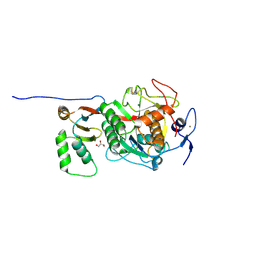 | | ProteinT protease, inactive mutant S224A | | Descriptor: | CALCIUM ION, Extracellular serine proteinase, GLYCEROL, ... | | Authors: | Nomme, J, Gavalda, S, Cioci, G, Guicherd, M, Marty, A, Duquesne, S, Ben Khaled, M. | | Deposit date: | 2023-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | An engineered enzyme embedded into PLA to make self-biodegradable plastic.
Nature, 631, 2024
|
|
8CAS
 
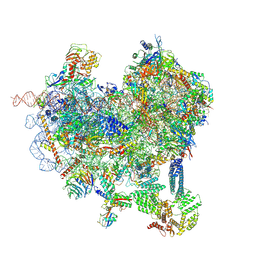 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8KB9
 
 | |
