8JN7
 
 | | Cryo-EM structure of the big tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C | | Descriptor: | Human antibody DENV-290 heavy chain, Human antibody DENV-290 light chain, Polyprotein (Fragment) | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8CQU
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1680) | | Descriptor: | CITRIC ACID, Putative NADH dehydrogenase/NAD(P)H nitroreductase, TERTIARY-BUTYL ALCOHOL | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQV
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_3392) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
5JPD
 
 | | Metal ABC transporter from Listeria monocytogenes with cadmium | | Descriptor: | CADMIUM ION, CHLORIDE ION, Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes with cadmium
to be published
|
|
5JPQ
 
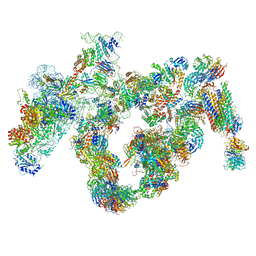 | | Cryo-EM structure of the 90S pre-ribosome | | Descriptor: | 18S ribosomal RNA, Bms1, Emg1, ... | | Authors: | Turk, M, Cheng, J, Berninghausen, O, Kornprobst, M, Flemming, D, Kos-Braun, I.C, Kos, M, Thoms, M, Hurt, E, Beckmann, R. | | Deposit date: | 2016-05-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome.
Cell, 166, 2016
|
|
8JN3
 
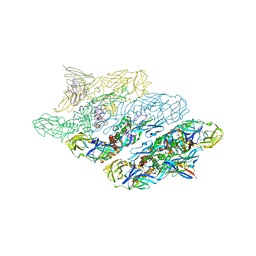 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 37 deg C (subparticle LLR-LRR) | | Descriptor: | Envelope protein, Human antibody DENV-115 heavy chain, Human antibody DENV-115 light chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8TNG
 
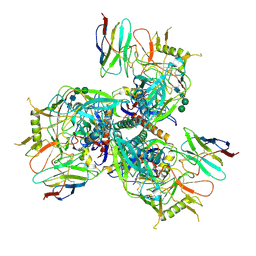 | |
8TNH
 
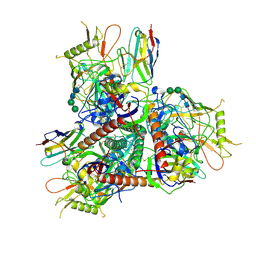 | |
8JN6
 
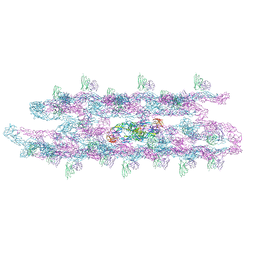 | | Cryo-EM structure of the small tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C | | Descriptor: | Human antibody DENV-290 heavy chain, Human antibody DENV-290 light chain, Polyprotein (Fragment) | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
5KBM
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), D113N Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
8DRS
 
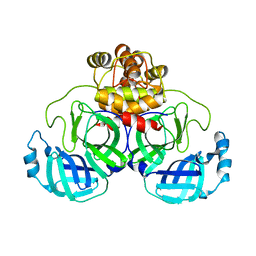 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
5JUW
 
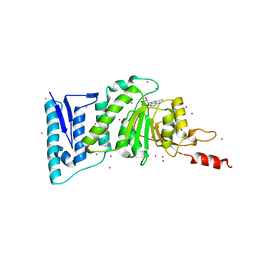 | | complex of Dot1l with SS148 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Li, Y, Spurr, S.S, Bayle, E.D, Fish, P.V, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complex of Dot1l with SS148
To Be Published
|
|
8TNI
 
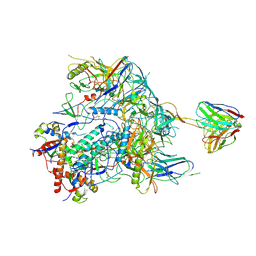 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex
To Be Published
|
|
7U8W
 
 | | hTRAP1 with inhibitors | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-fluoro-2H-isoindol-2-yl)methanone | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8X
 
 | | hTRAP1 with inhibitors | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-methoxy-2H-isoindol-2-yl)methanone | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8V
 
 | | hTRAP1 with inhibitors | | Descriptor: | (4-hydroxy-1,3-phenylene)bis[(2H-isoindol-2-yl)methanone], Heat shock protein 75 kDa, mitochondrial | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8U
 
 | | hTRAP1 with inhibitors | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, [2-hydroxy-5-(2H-isoindole-2-carbonyl)phenyl]{5-[3-(triphenyl-lambda~5~-phosphanyl)propoxy]-2H-isoindol-2-yl}methanone | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8K
 
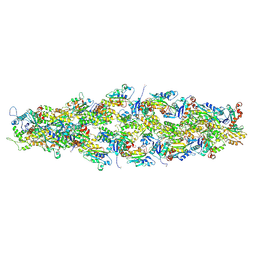 | | Magic Angle Spinning NMR Structure of Human Cofilin-2 Assembled on Actin Filaments | | Descriptor: | Actin, alpha skeletal muscle, Cofilin-2 | | Authors: | Kraus, J, Russell, R, Kudryashova, E, Xu, C, Katyal, N, Kudryashov, D, Perilla, J.R, Polenova, T. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | SOLID-STATE NMR | | Cite: | Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode.
Nat Commun, 13, 2022
|
|
5KEJ
 
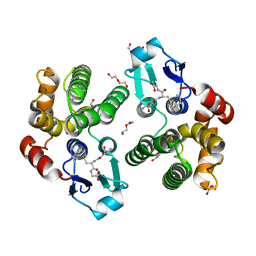 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with S-hexyl-glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-HEXYLGLUTATHIONE, Tau class glutathione S-transferase | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-06-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics.
Biochimie, 135, 2017
|
|
5JWE
 
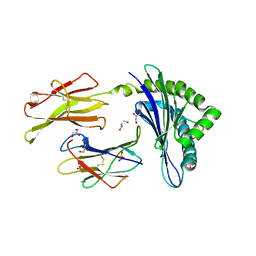 | | Crystal structure of H-2Db in complex with the LCMV-derived GP92-101 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
8DPD
 
 | |
8DRT
 
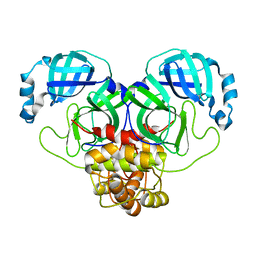 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14SQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
