5YMY
 
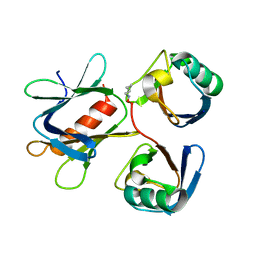 | | The structure of the complex between Rpn13 and K48-diUb | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Liu, Z, Dong, X, Gong, Z, Yi, H.W, Liu, K, Yang, J, Zhang, W.P, Tang, C. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-13 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13.
Cell Discov, 5, 2019
|
|
8CGV
 
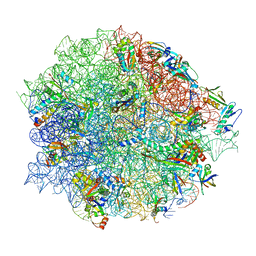 | | Tiamulin bound to the 50S subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.66 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5YOA
 
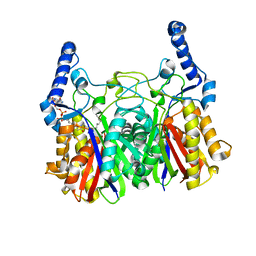 | |
5YTF
 
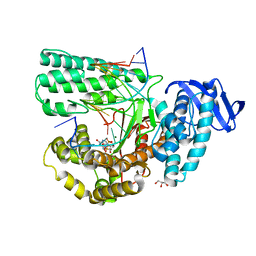 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4C38
 
 | | PKA-S6K1 Chimera with compound 21e (CCT239066) bound | | Descriptor: | 4-(1-ethyl-6-methyl-imidazo[4,5-c]pyridin-2-yl)-1,2,5-oxadiazol-3-amine, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR PEPTIDE, ... | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
6ZUF
 
 | | Urea-based Foldamer Inhibitor chimera C2 in complex with ASF1 Histone chaperone | | Descriptor: | C2 foldamer/peptide hybrid inhibitor of histone chaperone ASF1, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Mbianda, J, Perrin, E.M, Guerois, R, Legrand, P, Traore, S, Douat, C, Guichard, G, Ochsenbein, F. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Optimal anchoring of a foldamer inhibitor of ASF1 histone chaperone through backbone plasticity.
Sci Adv, 7, 2021
|
|
5YOL
 
 | | Crystal structure of octameric form of Nucleoside diphosphate kinase from Acinetobacter baumannii at 2.2 A resolution | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Singh, P.K, Sikarwar, J, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of octameric form of Nucleoside diphosphate kinase from Acinetobacter baumannii at 2.2 A resolution
To Be Published
|
|
8HRL
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
2GNQ
 
 | | Structure of wdr5 | | Descriptor: | CHLORIDE ION, WD-repeat protein 5 | | Authors: | Min, J, Schuetz, A, Allali-Hassani, A, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-10 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
8HRJ
 
 | |
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
2P6G
 
 | |
8GZD
 
 | |
7D99
 
 | | human potassium-chloride co-transporter KCC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
5YK7
 
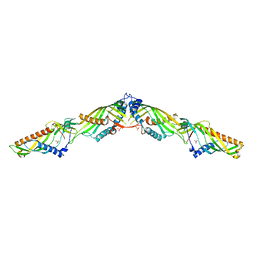 | | Crystal Structure of Mdm12-Mmm1 complex | | Descriptor: | Maintenance of mitochondrial morphology protein 1, Mitochondrial distribution and morphology protein 12, PHOSPHATE ION | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.799 Å) | | Cite: | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ZOP
 
 | | Structure of the cysteine-rich domain of PiggyMac, a domesticated PiggyBac transposase involved in programmed genome rearrangements | | Descriptor: | DDE_Tnp_1_7 domain-containing protein, ZINC ION | | Authors: | Bessa, L, Guerineau, M, Moriau, S, Lescop, E, Bontems, F, Mathy, N, Guittet, E, Bischerour, J, Betermier, M, Morellet, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of the PiggyMac cysteine-rich domain reveals zinc finger diversity in PiggyBac-related transposases.
Mob DNA, 12, 2021
|
|
6R7W
 
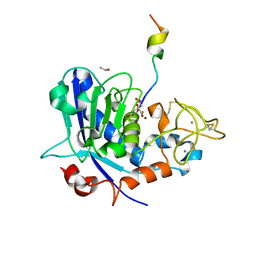 | | Tannerella forsythia mature mirolysin in complex with a cleaved peptide. | | Descriptor: | CALCIUM ION, CITRIC ACID, ETHANOL, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
8COL
 
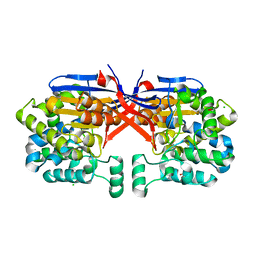 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (orthorombic form R4oP-2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imioloczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
8CLZ
 
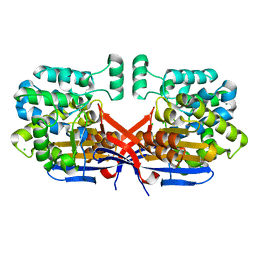 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (monoclinic form R4mC-2) | | Descriptor: | CHLORIDE ION, Putative L-asparaginase II protein, ZINC ION | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3BC3
 
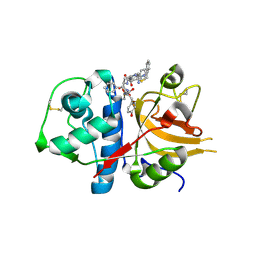 | | Exploring inhibitor binding at the S subsites of cathepsin L | | Descriptor: | Cathepsin L heavy and light chains, S-benzyl-N-(biphenyl-4-ylacetyl)-L-cysteinyl-N~5~-(diaminomethyl)-D-ornithyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Chowdhury, S.F, Joseph, L, Kumar, S, Tulsidas, S.R, Bhat, S, Ziomek, E, Nard, R.M, Sivaraman, J, Purisima, E.O. | | Deposit date: | 2007-11-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring inhibitor binding at the S' subsites of cathepsin L
J.Med.Chem., 51, 2008
|
|
8CLY
 
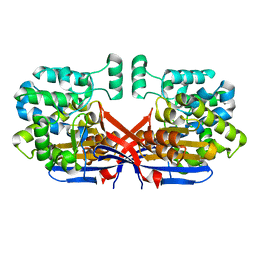 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (tetragonal form R4tP) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative L-asparaginase II protein, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8C8U
 
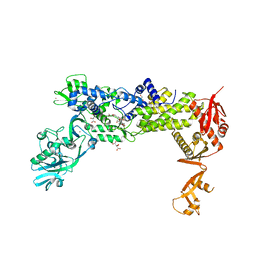 | | Priestia megaterium mupirocin-resistant isoleucyl-tRNA synthetase 2 complexed with mupirocin | | Descriptor: | Isoleucine--tRNA ligase, L(+)-TARTARIC ACID, MUPIROCIN, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Gruic-Sovulj, I, Ban, N. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
5YOX
 
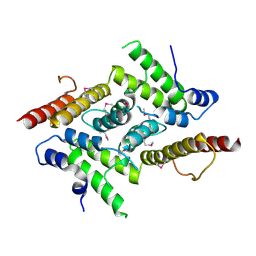 | | HD domain-containing protein YGK1(YGL101W) | | Descriptor: | HD domain-containing protein YGL101W, ZINC ION | | Authors: | Yang, J, Wang, F, Gao, Z, Zhou, K, Liu, Q. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | HD domain-containing protein YGK1(YGL101W)
To Be Published
|
|
6R3A
 
 | | BACTERIOPHAGE SPP1 MATURE CAPSID PROTEIN | | Descriptor: | Major capsid protein | | Authors: | Ignatiou, A, El Sadek Fadel, M, Buerger, J, Mielke, T, Topf, M, Tavares, P. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural transitions during the scaffolding-driven assembly of a viral capsid.
Nat Commun, 10, 2019
|
|
