6EOA
 
 | | Crystal Structure of HAL3 from Cryptococcus neoformans | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phosphopantothenoylcysteine decarboxylase | | Authors: | Reverter, D, Zhang, C, Molero, C, Arino, J. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Characterization of the atypical Ppz/Hal3 phosphatase system from the pathogenic fungus Cryptococcus neoformans.
Mol.Microbiol., 111, 2019
|
|
5N6L
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt C387A mutant from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5QGH
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000420a | | Descriptor: | (2R)-N-(1,2-oxazol-3-yl)-2-phenylbutanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5FJA
 
 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FKB
 
 | |
5FKW
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5QGR
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000022a | | Descriptor: | 5-(pyridin-2-yl)thiophene-2-carbothioamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5N8P
 
 | |
1HAI
 
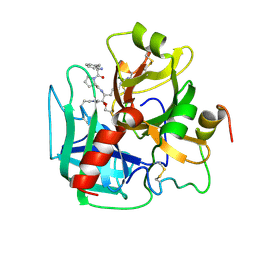 | | THE ISOMORPHOUS STRUCTURES OF PRETHROMBIN2, HIRUGEN-AND PPACK-THROMBIN: CHANGES ACCOMPANYING ACTIVATION AND EXOSITE BINDING TO THROMBIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Vijayalakshmi, J. | | Deposit date: | 1994-06-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin.
Protein Sci., 3, 1994
|
|
5QH5
 
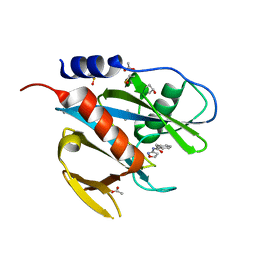 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000224a | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, N-(1,2-oxazol-3-yl)-2-phenylacetamide, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5FO3
 
 | | ZapC cell division regulator from E. coli | | Descriptor: | CELL DIVISION PROTEIN ZAPC | | Authors: | Ortiz, C, Kureisaite-Ciziene, D, Schmitz, F, Vicente, M, Lowe, J. | | Deposit date: | 2015-11-18 | | Release date: | 2015-11-25 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Z-Ring Associated Cell Division Protein Zapc from Escherichia Coli.
FEBS Lett., 589, 2015
|
|
5FOE
 
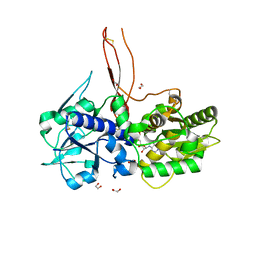 | | Crystal structure of the C. elegans Protein O-fucosyltransferase 2 (CePOFUT2) double mutant (R298K-R299K) in complex with GDP and the human TSR1 from thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 2,Thrombospondin-1, ... | | Authors: | Valero-Gonzalez, J, Leonhard-Melief, C, Lira-Navarrete, E, Jimenez-Oses, G, Hernandez-Ruiz, C, Pallares, M.C, Yruela, I, Vasudevan, D, Lostao, A, Corzana, F, Takeuchi, H, Haltiwanger, R.S, Hurtado-Guerrero, R. | | Deposit date: | 2015-11-19 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Proactive Role of Water Molecules in Acceptor Recognition of Thrombospondin Type 1 Repeats by Protein-O-Fucosyltransferase 2
Nat.Chem.Biol., 12, 2016
|
|
5SZD
 
 | | Crystal structure of Aquifex aeolicus Hfq at 1.5A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Stanek, K, Patterson, J, Randolph, P.S, Mura, C. | | Deposit date: | 2016-08-13 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NJG
 
 | | Structure of an ABC transporter: part of the structure that could be built de novo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
1H8D
 
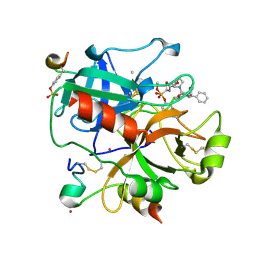 | | X-ray structure of the human alpha-thrombin complex with a tripeptide phosphonate inhibitor. | | Descriptor: | HIRUDIN I, N-[(benzyloxy)carbonyl]-beta-phenyl-D-phenylalanyl-N-{(1S,3E)-1-[dihydroxy(diphenoxy)-lambda~5~-phosphanyl]-4-methoxybut-3-en-1-yl}-L-prolinamide, THROMBIN | | Authors: | Skordalakes, E, Dodson, G.G, Green, D, Deadman, J. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of Human Alpha-Thrombin by a Phosphonate Tripeptide Proceeds Via a Metastable Pentacoordinated Phosphorus Intermediate
J.Mol.Biol., 311, 2001
|
|
5FNU
 
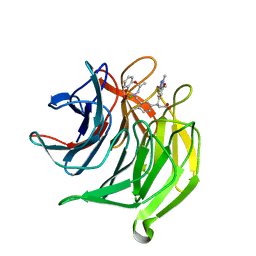 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
6E8G
 
 | |
5NBD
 
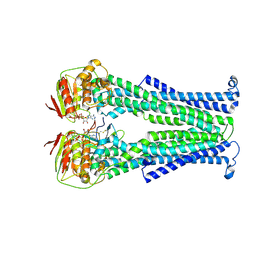 | | PglK flippase in complex with inhibitory nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nanobody, WlaB protein | | Authors: | Perez, C, Pardon, E, Steyaert, J, Locher, K.P. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Sci Rep, 7, 2017
|
|
6EE8
 
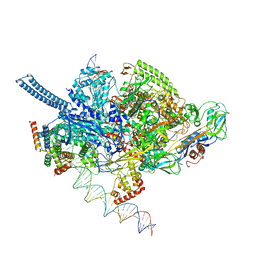 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (60-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
5FO5
 
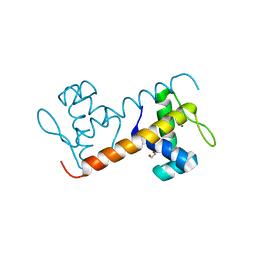 | | Structure of the DNA-binding domain of Escherichia coli methionine biosynthesis regulator MetR | | Descriptor: | 1,2-ETHANEDIOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR METR, MAGNESIUM ION | | Authors: | Punekar, A.S, Porter, J, Urbanowski, M.L, Stauffer, G.V, Carr, S.B, Phillips, S.E. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for DNA Recognition by the Transcription Regulator Metr.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5NEF
 
 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with guanidine | | Descriptor: | GUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5N02
 
 | | Crystal structure of the decarboxylase AibA/AibB C56S variant | | Descriptor: | ACETATE ION, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5QGO
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with OX-220 | | Descriptor: | (3aR,4R,6R,7R,8aR)-6-phenyloctahydro-1H-3a,7-epiminocyclohepta[c]pyrrol-4-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5NOG
 
 | | Ca2+-induced Movement of Tropomyosin on Native Cardiac Thin Filaments - "Blocked" state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cardiac muscle alpha actin 1, MAGNESIUM ION, ... | | Authors: | Risi, C, Eisner, J, Belknap, B, Heeley, D.H, White, H.D, Schroeder, G.F, Galkin, V.E. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-19 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5QGZ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000161 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-2-(4-fluorophenyl)acetamide, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
