8QJP
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with uridine - 5'- monophosphate | | Descriptor: | GLYCEROL, PHOSPHATE ION, S1/P1 Nuclease, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of the Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study
The FEBS Journal, 2024
|
|
6FUI
 
 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | (1~{R},2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]cyclohexane-1-carboxylic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6SSP
 
 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a NAM nanobody | | Descriptor: | CALCIUM ION, Cys-loop ligand-gated ion channel, NANOBODY 21, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
6SV6
 
 | | Non-terahertz irradiated structure of bovine trypsin (even frames of crystal x38) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-17 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
2CVY
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | 9-per peptide from Ribonucleoside-diphosphate reductase small chain 1, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1, ... | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6SVN
 
 | | Reference structure of bovine trypsin (even frames of crystal x28) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SW5
 
 | | Crystal structure of the human S-adenosylmethionine synthetase 1 (ligand-free form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, S-adenosylmethionine synthase isoform type-1 | | Authors: | Panmanee, J, Antoyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the dominant inheritance of hypermethioninemia associated with the Arg264His mutation in the MAT1A gene.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6SX6
 
 | | Guanine-rich oligonucleotide with 5'-GC end form G-quadruplex with A(GGGG)A hexad, GCGC- and G-quartets and two symmetric GG and AA base pairs | | Descriptor: | GCn | | Authors: | Pavc, D, Wang, B, Spindler, L, Drevensek-Olenik, I, Plavec, J, Sket, P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly.
Nucleic Acids Res., 48, 2020
|
|
7B3U
 
 | | OXA-10 beta-lactamase with covalent modification | | Descriptor: | Beta-lactamase OXA-10, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-12-01 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6G13
 
 | | C-terminal domain of MERS-CoV nucleocapsid | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Nguyen, T.H.V, Ferron, F.P, Lichiere, J, Canard, B, Papageorgiou, N, Coutard, B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7B3S
 
 | | OXA-10 beta-lactamase with S67Dha modification | | Descriptor: | Beta-lactamase OXA-10, CARBON DIOXIDE, SODIUM ION, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-12-01 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4GQM
 
 | |
7B3R
 
 | |
6G44
 
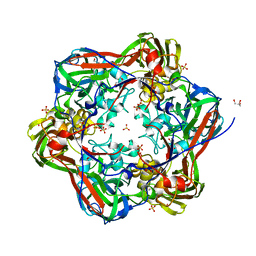 | | Crystal structure of mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | GLYCEROL, Putative major capsid protein, SULFATE ION | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1P5V
 
 | | X-ray structure of the Caf1M:Caf1 chaperone:subunit preassembly complex | | Descriptor: | Chaperone protein Caf1M, F1 capsule antigen | | Authors: | Zavialov, A.V, Berglund, J, Pudney, A.F, Fooks, L.J, Ibrahim, T.M, MacIntyre, S, Knight, S.D. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-24 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Biogenesis of the Capsular F1 Antigen from Yersinia pestis. Preserved Folding Energy Drives Fiber Formation
Cell(Cambridge,Mass.), 113, 2003
|
|
2JGA
 
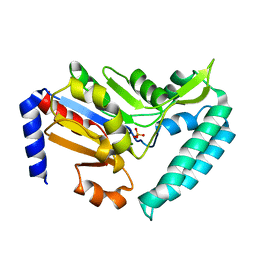 | | Crystal structure of human cytosolic 5'-nucleotidase III in complex with phosphate and magnesium | | Descriptor: | CYTOSOLIC 5'-NUCLEOTIDASE III, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wallden, K, Stenmark, P, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, M, Holmberg, B, Schiavone, L, Hogbom, M, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2007-02-09 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Human Cytosolic 5'-Nucleotidase II: Insights Into Allosteric Regulation and Substrate Recognition.
J.Biol.Chem., 282, 2007
|
|
6G53
 
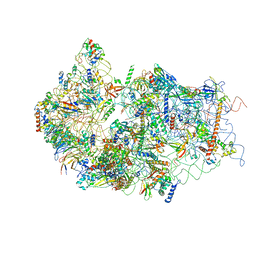 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State E | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
5OVI
 
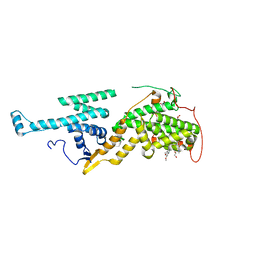 | | Ras guanine nucleotide exchange factor SOS1 (Rem-cdc25) in complex with small molecule inhibitor BAY-293 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-2-methyl-~{N}-[(1~{R})-1-[4-[2-(methylaminomethyl)phenyl]thiophen-2-yl]ethyl]quinazolin-4-amine, Son of sevenless homolog 1 | | Authors: | Hillig, R.C, Sautier, B, Schroeder, J, Moosmayer, D, Hilpmann, A, Stegmann, C.M, Briem, H, Boemer, U, Weiske, J, Badock, V, Petersen, K, Kahmann, J, Wegener, D, Bohnke, N, Eis, K, Graham, K, Wortmann, L, von Nussbaum, F, Bader, B. | | Deposit date: | 2017-08-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6SFT
 
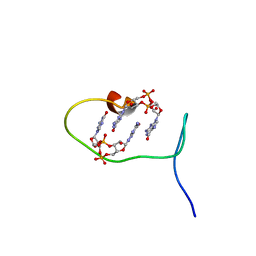 | | Solution structure of protein ARR_CleD in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Two-component receiver protein CleD | | Authors: | Habazettl, J, Hee, C.S, Jenal, U, Schirmer, T, Grzesiek, S. | | Deposit date: | 2019-08-02 | | Release date: | 2020-06-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6G43
 
 | | Crystal structure of SeMet-labeled mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | Putative major capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6SNH
 
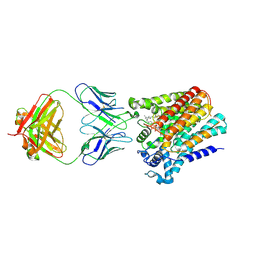 | | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc | | Descriptor: | 6AG9 Fab heavy chain, 6AG9 Fab light chain, Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
7W5H
 
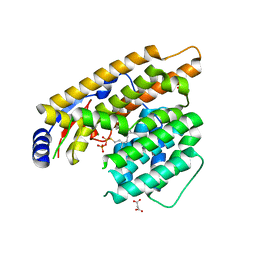 | | The structure of trichobrasilenol synthase TaTC6 in complex with FsPP | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
2J8Q
 
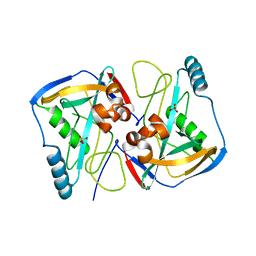 | | Crystal structure of human cleavage and polyadenylation specificity factor 5 (CPSF5) in complex with a sulphate ion. | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 5, SULFATE ION | | Authors: | Moche, M, Stenmark, P, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg, S.L, Hogbom, M, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Upsten, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of Human Cleavage and Polyadenylation Specific Factor-5 Reveals a Dimeric Nudix Protein with a Conserved Catalytic Site.
Proteins, 73, 2008
|
|
2CVW
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1, ... | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CW9
 
 | | Crystal structure of human Tim44 C-terminal domain | | Descriptor: | PENTAETHYLENE GLYCOL, translocase of inner mitochondrial membrane | | Authors: | Handa, N, Kishishita, S, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Uchikubo, T, Takemoto, C, Jin, Z, Chrzas, J, Chen, L, Liu, Z.-J, Wang, B.-C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
